Authors
This author initiated 26 original pathways:
This author contributed to 627 pathways:
This author contributed to 627 pathways:
AlexanderPico
Alex Pico - Gladstone Institutes, San Francisco, CA, USA
He co-founded WikiPathways in 2007, as an open, collaborative platform for molecular pathway curation. In 2010, he took on leadership roles in the Cytoscape Consortium as vice president and as executive director for the National Resource for Network Biology. In addition to his independent research, he has served as the director of the Gladstone Bioinformatics Core since 2020.
https://gladstone.org/people/alex-pico
- GitHub: AlexanderPico
- ORCiD: 0000-0001-5706-2163
- Linked In: apico
- Google Scholar: FUsTMX8AAAAJ
- Wikidata: Q28530149
- Twitter: xanderpico
- Mastodon: https://qoto.org/@xanderpico
- Email: alex.pico@gladstone.ucsf.edu
Communities
COVID-19 CPTACPathways
This author initiated 26 original pathways:
This author contributed to 627 pathways:
This author contributed to 627 pathways:
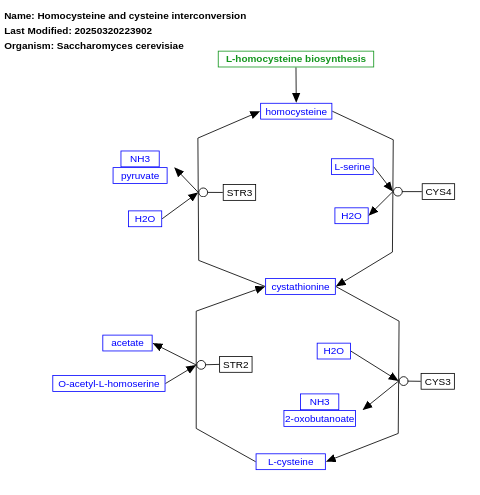
- Homocysteine and cysteine interconversion - WP128 (Saccharomyces cerevisiae)
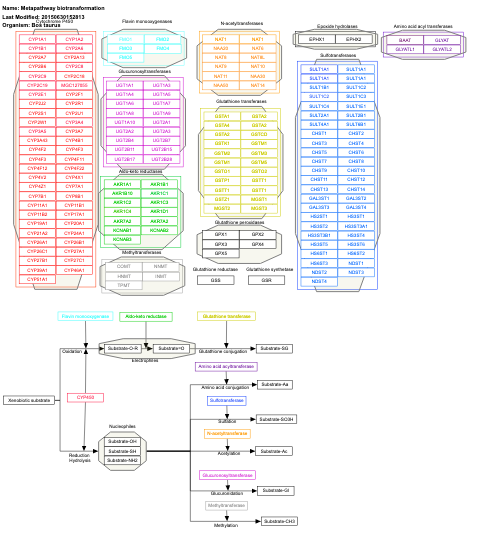
- Metapathway biotransformation - WP1006 (Bos taurus)
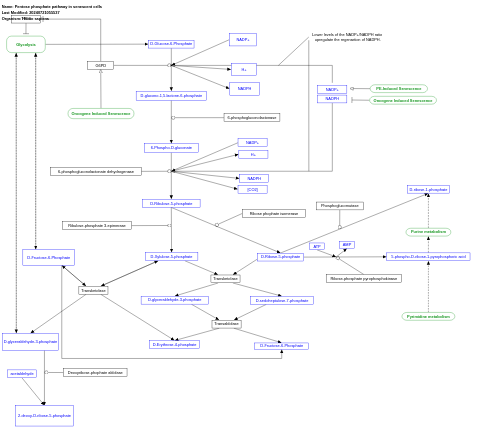
- Pentose phosphate pathway in senescent cells - WP5043 (Homo sapiens)
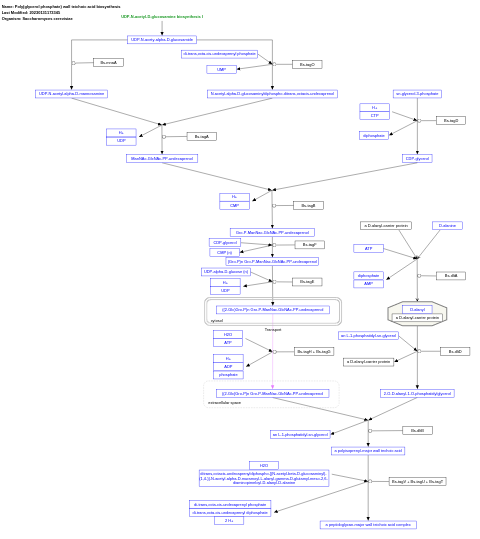
- Poly(glycerol phosphate) wall teichoic acid biosynthesis - WP4162 (Saccharomyces cerevisiae)
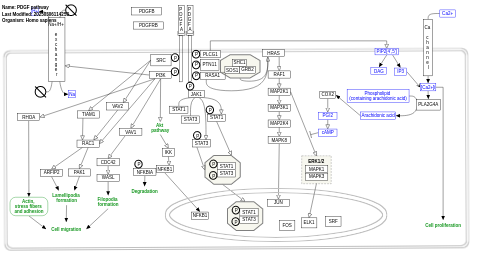
- PDGF pathway - WP2526 (Homo sapiens)
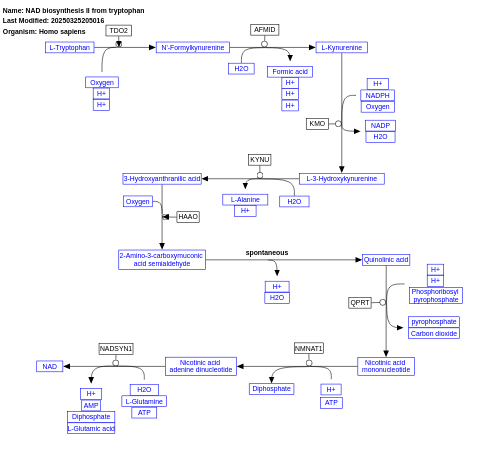
- NAD biosynthesis II from tryptophan - WP2485 (Homo sapiens)
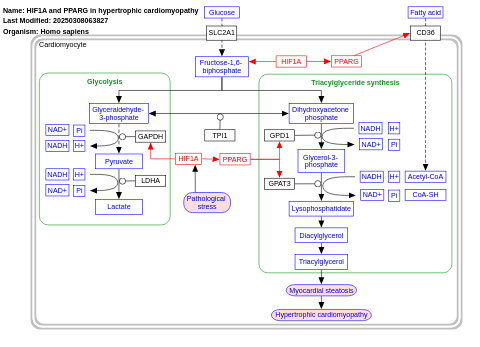
- HIF1A and PPARG in hypertrophic cardiomyopathy - WP2456 (Homo sapiens)
- Pantothenate and coenzyme A biosynthesis - WP462 (Saccharomyces cerevisiae)
- Aerobic glycolysis - augmented - WP4628 (Homo sapiens)
- Catalytic cycle of mammalian flavin-containing monooxygenases (FMOs) - WP688 (Homo sapiens)
- Threonine biosynthesis - WP331 (Saccharomyces cerevisiae)
- Deregulating cellular metabolism through non-genotoxic carcinogens - WP5571 (Homo sapiens)
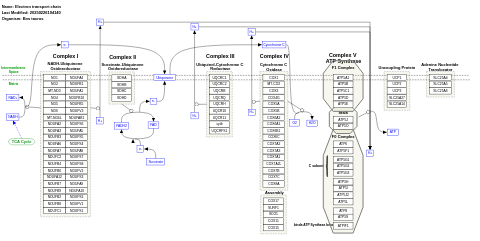
- Electron transport chain - WP1119 (Canis familiaris)
- 5q35 copy number variation - WP5380 (Homo sapiens)
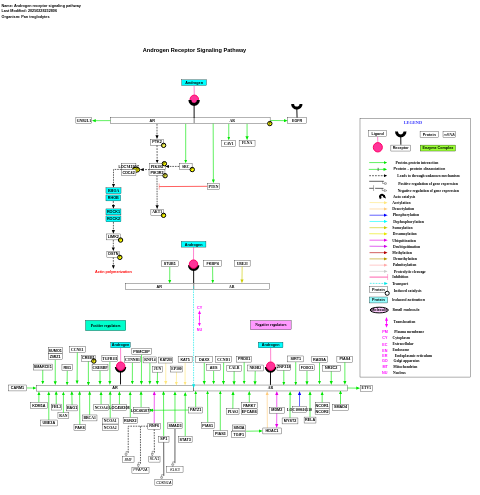
- Androgen receptor signaling pathway - WP252 (Mus musculus)
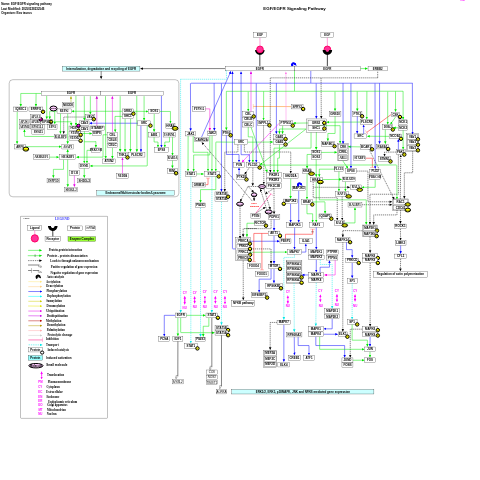
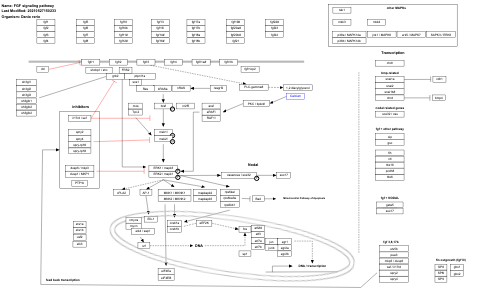
- EGFR1 signaling pathway - WP1323 (Danio rerio)
- Electron transport chain - WP1002 (Bos taurus)
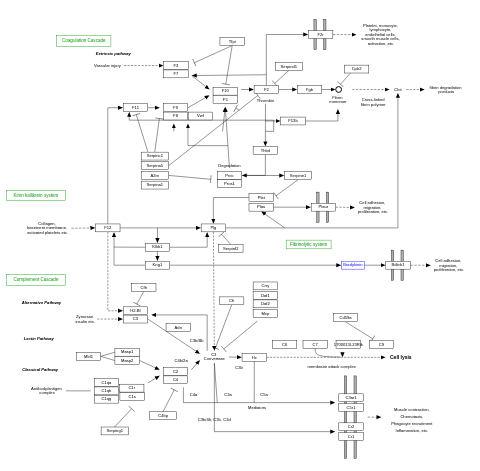
- Complement and coagulation cascades - WP449 (Mus musculus)
- B cell receptor signaling pathway - WP274 (Mus musculus)
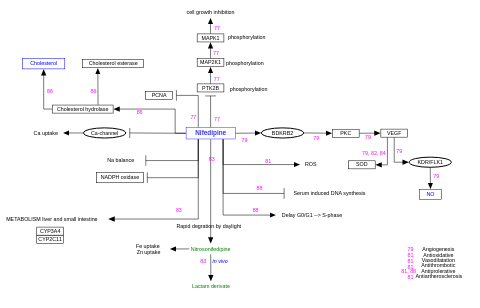
- Nifedipine activity - WP259 (Homo sapiens)
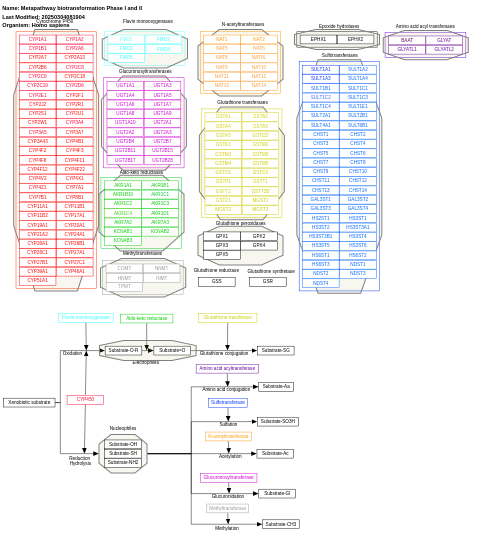
- Metapathway biotransformation Phase I and II - WP702 (Homo sapiens)
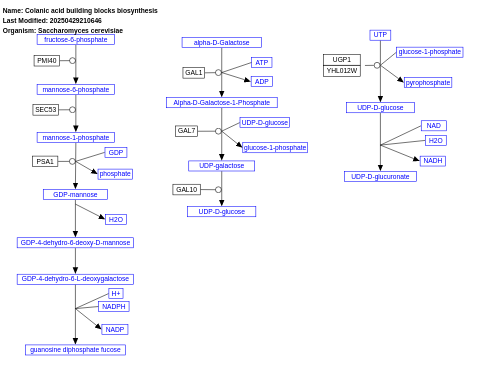
- Colanic acid building blocks biosynthesis - WP121 (Saccharomyces cerevisiae)
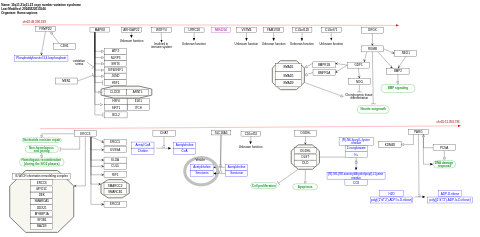
- 10q11.21q11.23 copy number variation syndrome - WP5352 (Homo sapiens)
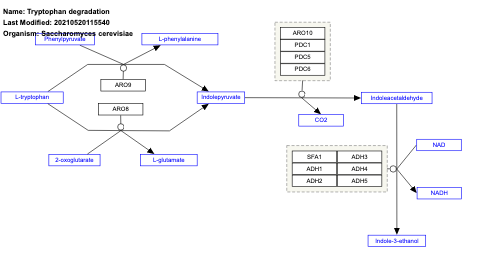
- Tryptophan degradation - WP301 (Saccharomyces cerevisiae)
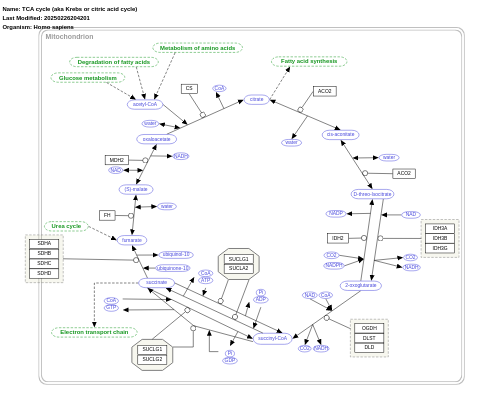
- TCA cycle (aka Krebs or citric acid cycle) - WP78 (Homo sapiens)
- ErbB signaling - WP673 (Homo sapiens)
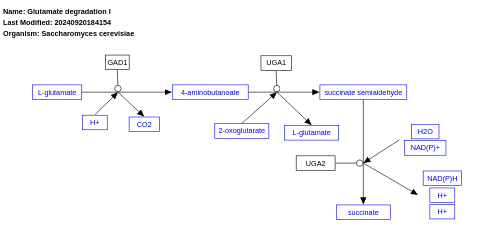
- Glutamate degradation I - WP556 (Saccharomyces cerevisiae)
- Clear cell renal cell carcinoma pathways - WP4018 (Homo sapiens)
- MAPK signaling - WP382 (Homo sapiens)
- Focal adhesion - WP306 (Homo sapiens)
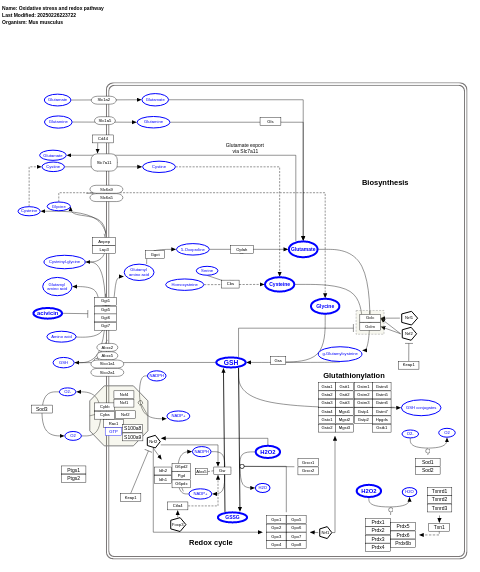
- Oxidative stress and redox pathway - WP4466 (Mus musculus)
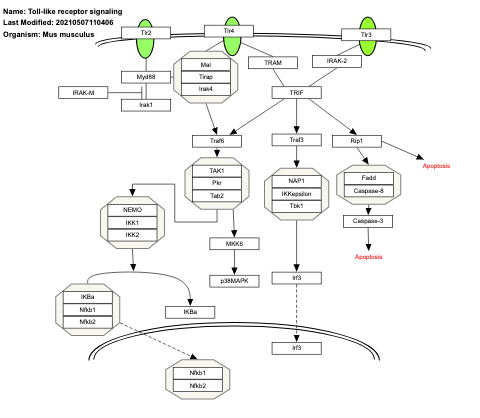
- Toll-like receptor signaling - WP75 (Homo sapiens)
- Pleural mesothelioma - WP5087 (Homo sapiens)
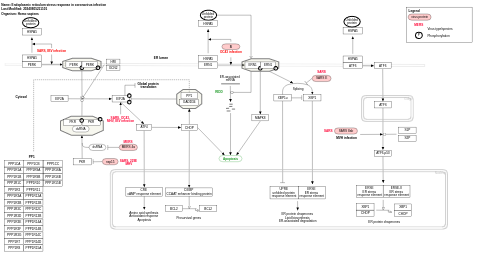
- Endoplasmic reticulum stress response in coronavirus infection - WP4861 (Homo sapiens)
- Alzheimer's disease and miRNA effects - WP2059 (Homo sapiens)
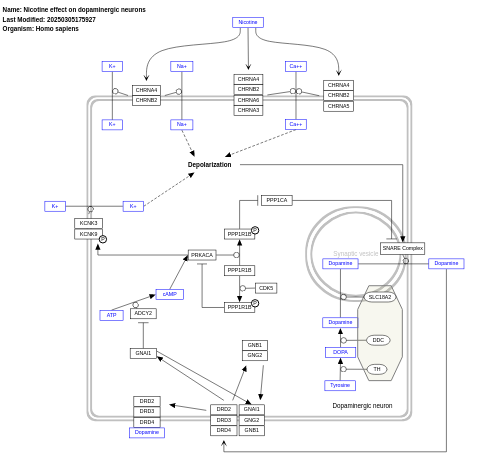
- Nicotine effect on dopaminergic neurons - WP1602 (Homo sapiens)
- Parthanatos cell death signaling pathway - WP5585 (Homo sapiens)
- Lactate shuttle in glial cells - WP5314 (Homo sapiens)
- Electron transport chain - WP295 (Mus musculus)
- Vitamin B12 metabolism - WP1533 (Homo sapiens)
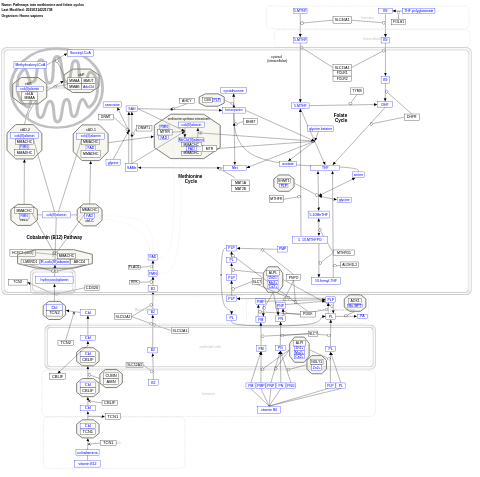
- Pathways into methionine and folate cycles - WP5488 (Homo sapiens)
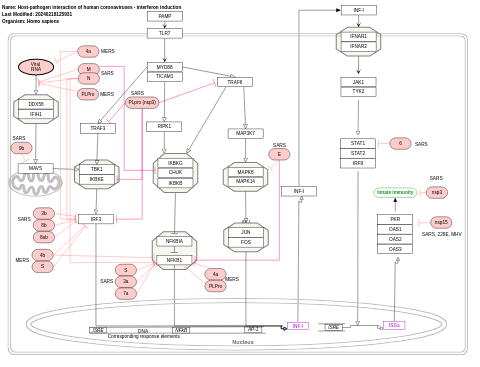
- Host-pathogen interaction of human coronaviruses - interferon induction - WP4880 (Homo sapiens)
- Folate metabolism - WP176 (Homo sapiens)
- Electron transport chain - WP1339 (Danio rerio)
- Folic acid network - WP1311 (Rattus norvegicus)
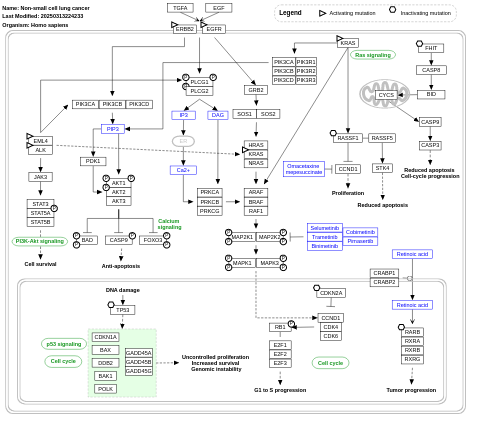
- Non-small cell lung cancer - WP4255 (Homo sapiens)
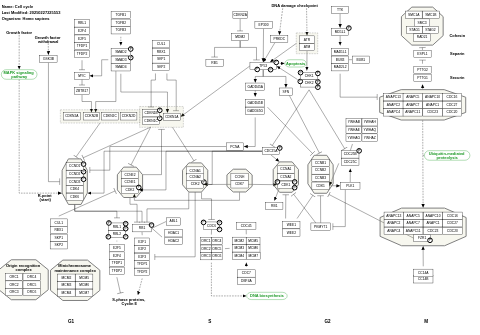
- Cell cycle - WP179 (Homo sapiens)
- Killer cell immunoglobulin-like receptors and human leukocyte antigen C pathway - WP5576 (Homo sapiens)
- Oligodendrocyte development - WP5574 (Homo sapiens)
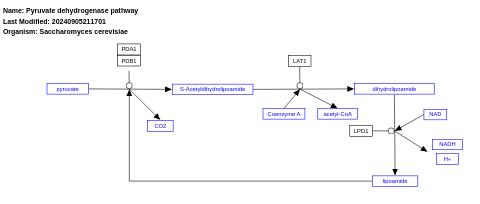
- Pyruvate dehydrogenase pathway - WP214 (Saccharomyces cerevisiae)
- Carbon assimilation C4 pathway - WP1493 (Zea mays)
- 8p23.1 copy number variation syndrome - WP5346 (Homo sapiens)
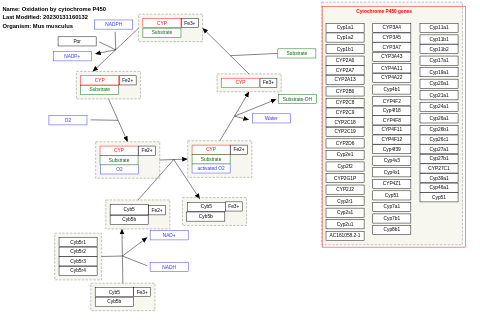
- Oxidation by cytochrome P450 - WP1274 (Mus musculus)
- Selenium micronutrient network - WP15 (Homo sapiens)
- Folic acid network - WP1273 (Mus musculus)
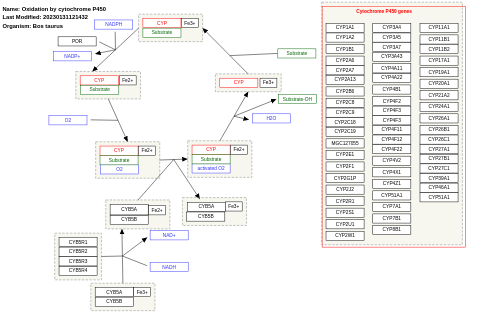
- Oxidation by cytochrome P450 - WP1077 (Bos taurus)
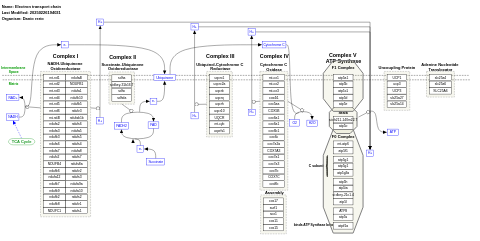
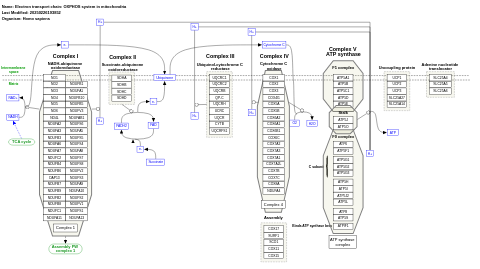
- Electron transport chain: OXPHOS system in mitochondria - WP111 (Homo sapiens)
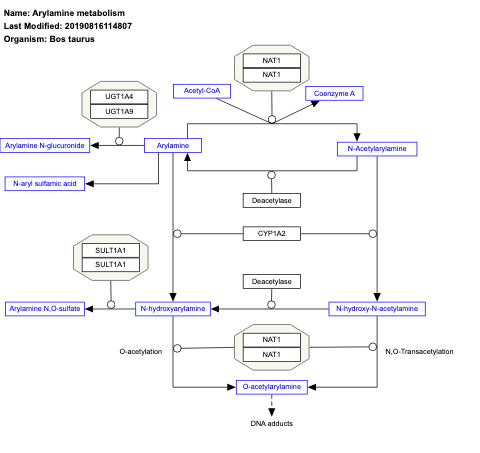
- Arylamine metabolism - WP993 (Bos taurus)
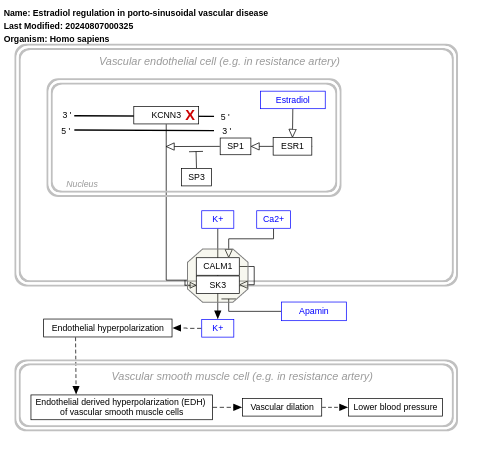
- Estradiol regulation in porto-sinusoidal vascular disease - WP5235 (Homo sapiens)
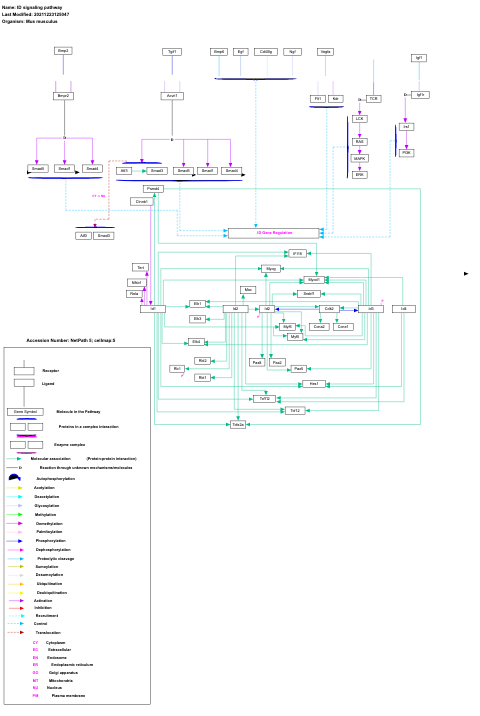
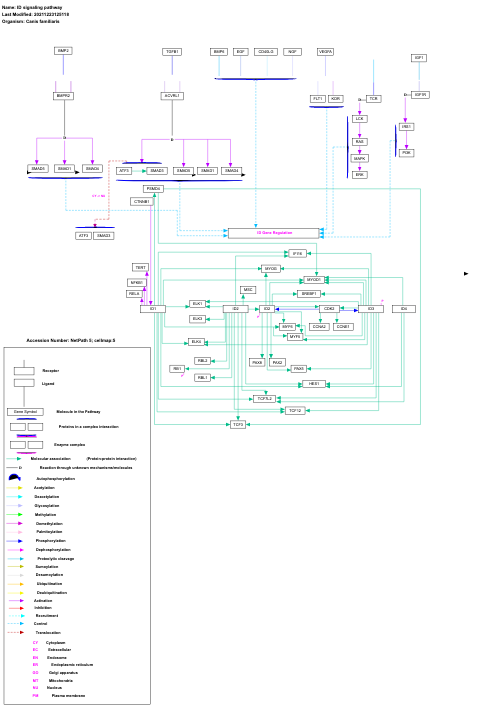
- ID signaling pathway - WP512 (Mus musculus)
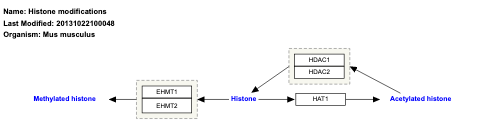
- Histone modifications - WP300 (Mus musculus)
- Regulation of toll-like receptor signaling - WP1449 (Homo sapiens)
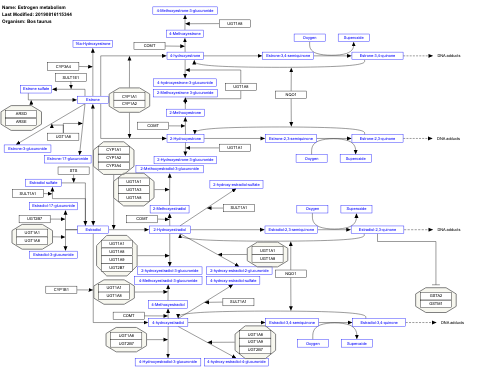
- Estrogen metabolism - WP1053 (Bos taurus)
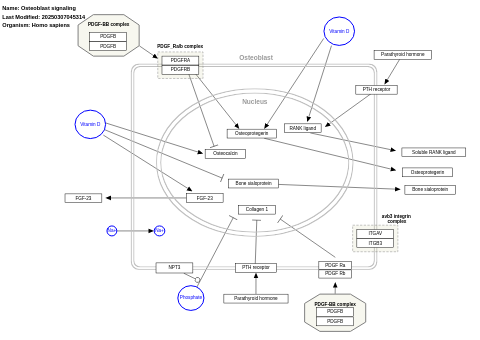
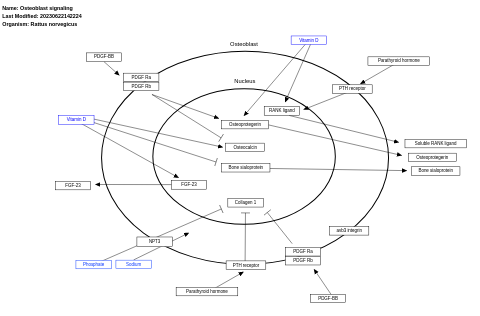
- Osteoblast signaling - WP322 (Homo sapiens)
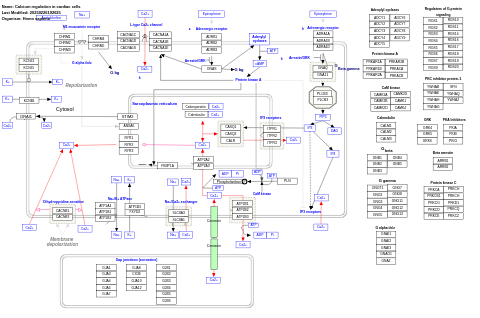
- Calcium regulation in cardiac cells - WP808 (Gallus gallus)
- SARS-CoV-2 and COVID-19 pathway - WP4846 (Homo sapiens)
- Ergosterol biosynthesis - WP5354 (Saccharomyces cerevisiae)
- Endothelin pathways - WP2197 (Homo sapiens)
- CCL18 signaling - WP5097 (Homo sapiens)
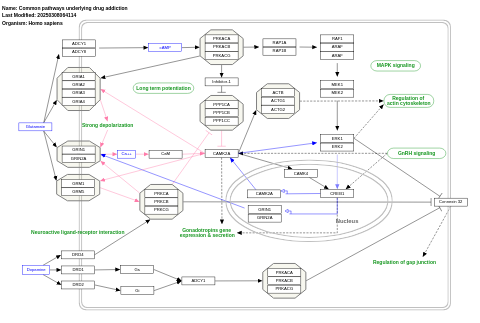
- Common pathways underlying drug addiction - WP2636 (Homo sapiens)
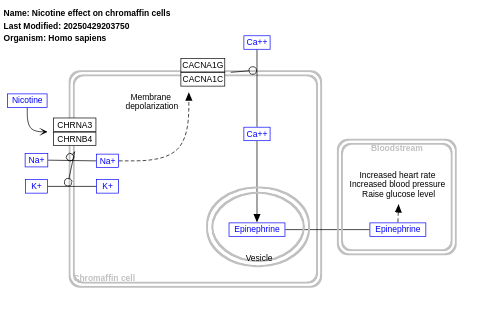
- Nicotine effect on chromaffin cells - WP1603 (Homo sapiens)
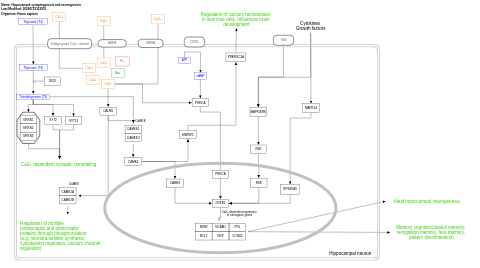
- Hippocampal synaptogenesis and neurogenesis - WP5231 (Homo sapiens)
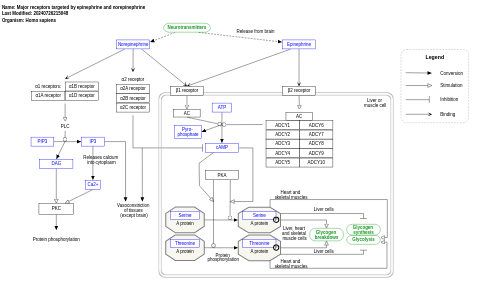
- Major receptors targeted by epinephrine and norepinephrine - WP4589 (Homo sapiens)
- Female steroid hormones in cardiomyocyte energy metabolism - WP5318 (Homo sapiens)
- Prader-Willi and Angelman syndrome - WP3998 (Homo sapiens)
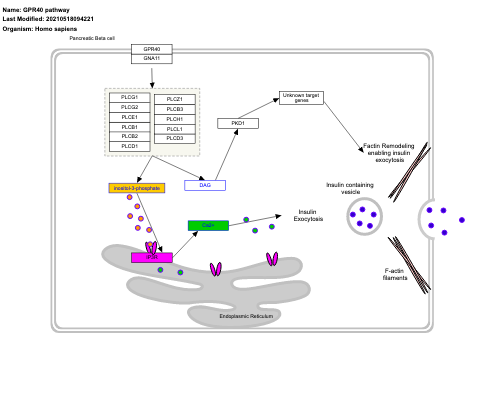
- GPR40 role in insulin secretion - WP3958 (Homo sapiens)
- Histone modifications - WP2369 (Homo sapiens)
- Chronic hyperglycemia impairment of neuron function - WP5283 (Homo sapiens)
- 7-oxo-C and 7-beta-HC pathways - WP5064 (Homo sapiens)
- Angiotensin II signaling (acute) in thick ascending limbs - WP3887 (Rattus norvegicus)
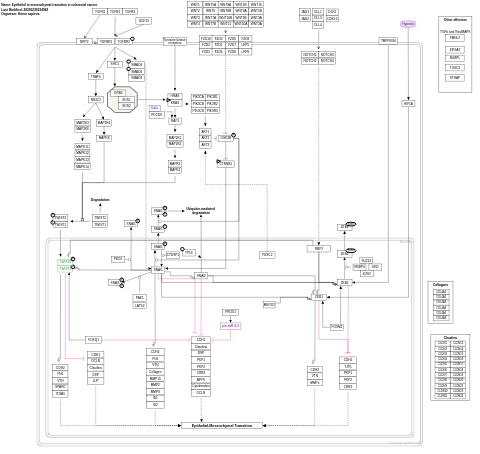
- Epithelial to mesenchymal transition in colorectal cancer - WP4239 (Homo sapiens)
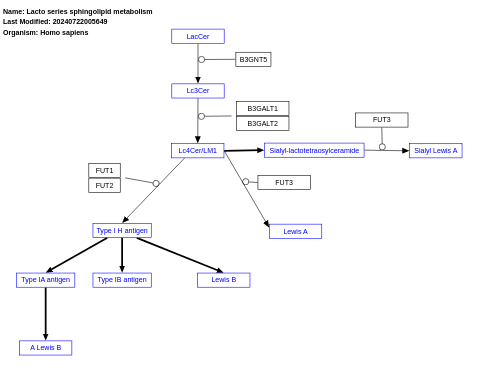
- Lacto series sphingolipid metabolism - WP5303 (Homo sapiens)
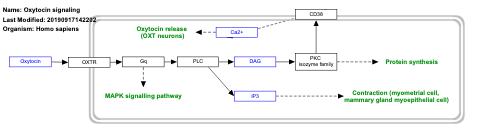
- Oxytocin signaling - WP2889 (Homo sapiens)
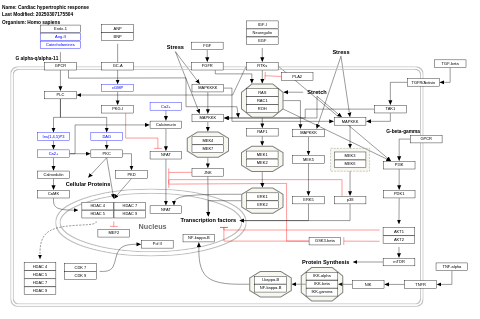
- Cardiac hypertrophic response - WP2795 (Homo sapiens)
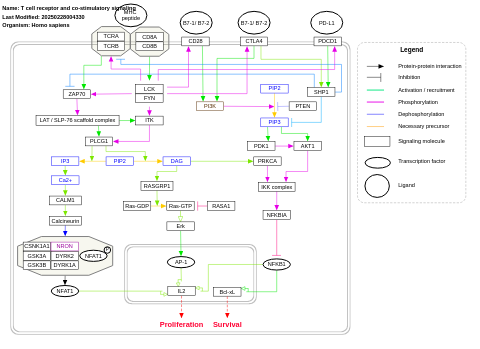
- T cell receptor and co-stimulatory signaling - WP2583 (Homo sapiens)
- Amyotrophic lateral sclerosis (ALS) - WP2447 (Homo sapiens)
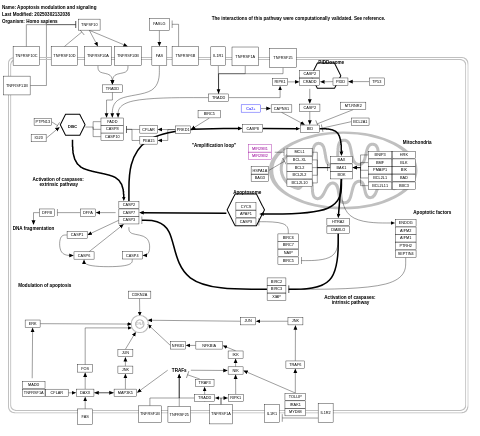
- Apoptosis modulation and signaling - WP1772 (Homo sapiens)
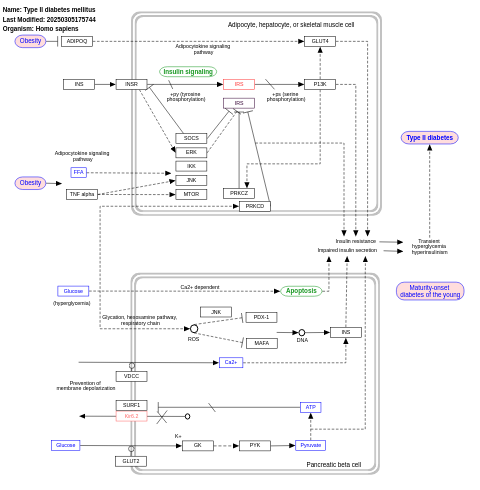
- Type II diabetes mellitus - WP1584 (Homo sapiens)
- Thyroxine (thyroid hormone) production - WP1981 (Homo sapiens)
- N-glycan biosynthesis - WP5196 (Ovis aries)
- Osteoclast signaling - WP890 (Pan troglodytes)
- ACE inhibitor pathway - WP554 (Homo sapiens)
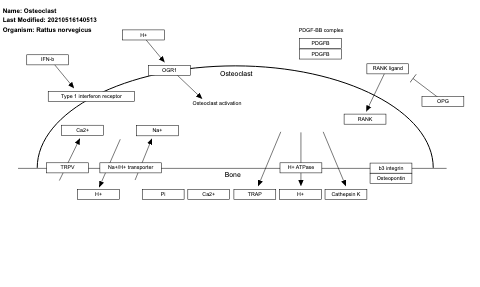
- Osteoclast - WP489 (Rattus norvegicus)
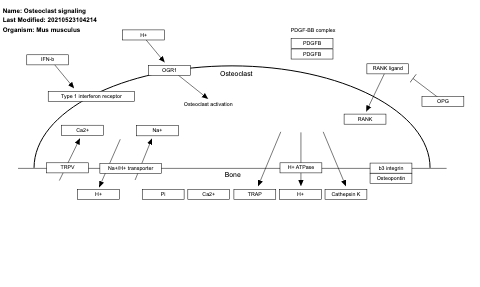
- Osteoclast signaling - WP454 (Mus musculus)
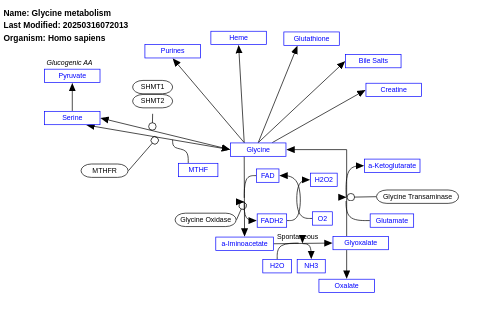
- Glycine metabolism - WP1495 (Homo sapiens)
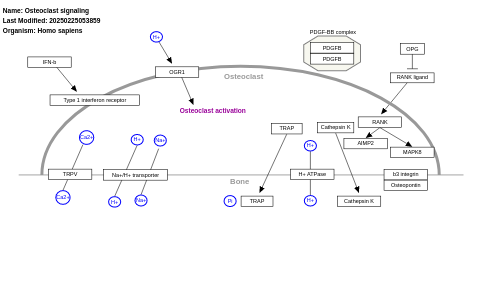
- Osteoclast signaling - WP12 (Homo sapiens)
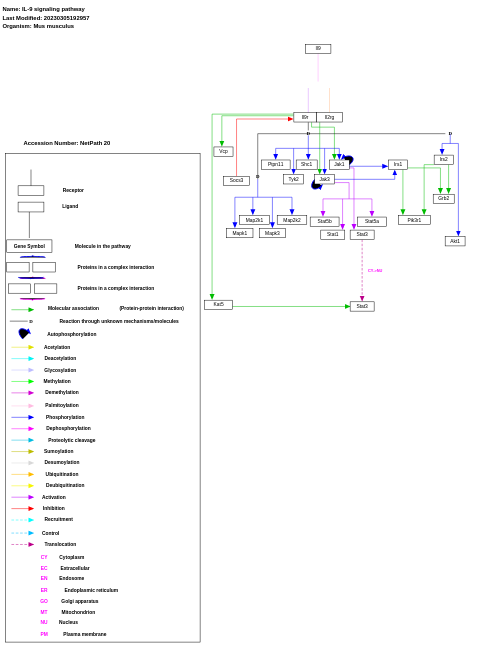
- IL-9 signaling pathway - WP10 (Mus musculus)
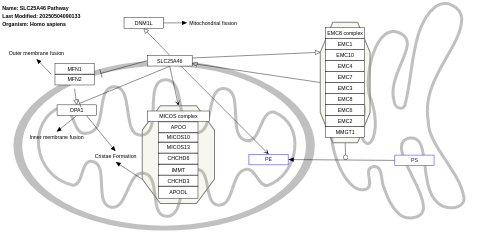
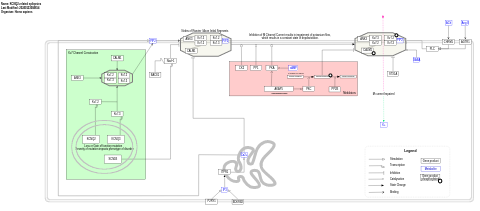
- SLC25A46 pathway - WP5521 (Homo sapiens)
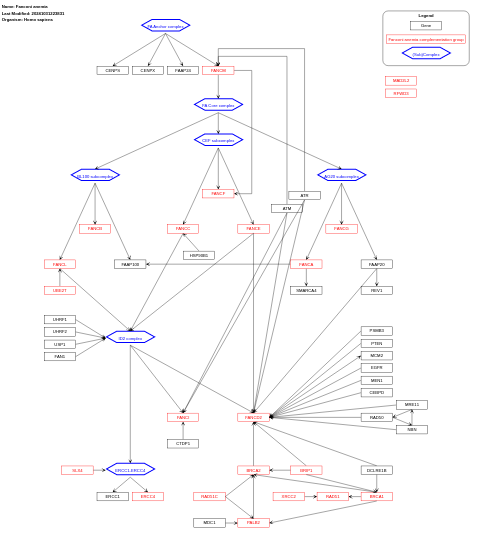
- Fanconi anemia - WP5465 (Homo sapiens)
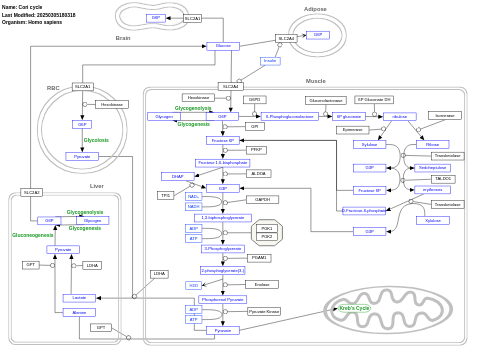
- Cori cycle - WP1946 (Homo sapiens)
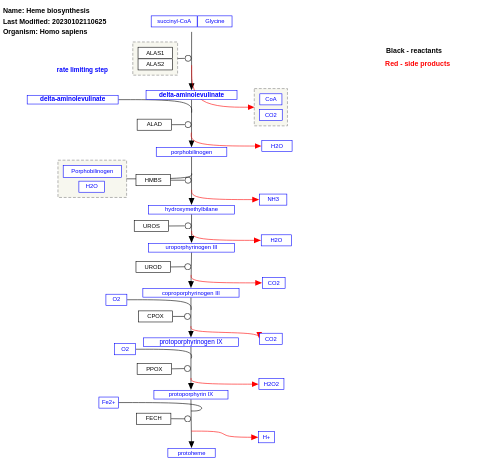
- Heme biosynthesis - WP561 (Homo sapiens)
- Neural crest cell migration in cancer - WP4565 (Homo sapiens)
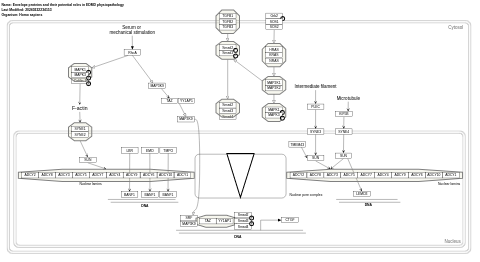
- Envelope proteins and their potential roles in EDMD physiopathology - WP4535 (Homo sapiens)
- KCNQ2-related epilepsies - WP5360 (Homo sapiens)
- Calcium regulation in cardiac cells - WP536 (Homo sapiens)
- Affected pathways in Duchenne muscular dystrophy - WP5356 (Homo sapiens)
- Kallmann syndrome - WP5074 (Homo sapiens)
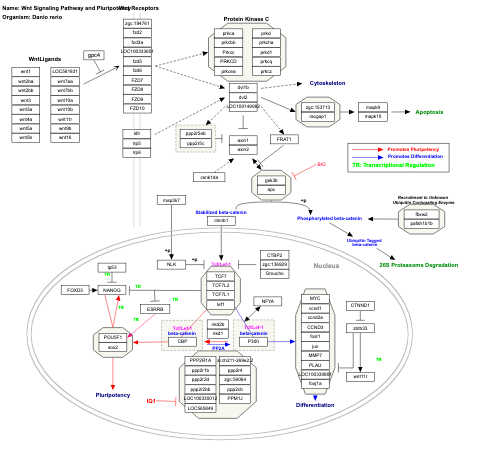
- Wnt signaling - WP428 (Homo sapiens)
- G protein signaling - WP35 (Homo sapiens)
- ESC pluripotency pathways - WP339 (Mus musculus)
- Non-canonical Wnt pathway - WP215 (Danio rerio)
- BDNF-TrkB signaling - WP3676 (Homo sapiens)
- Angiotensin II receptor type 1 pathway - WP5036 (Homo sapiens)
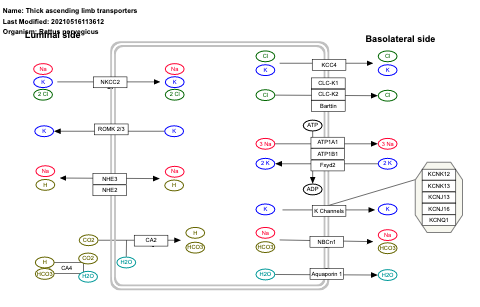
- Thick ascending limb transporters - WP3882 (Rattus norvegicus)
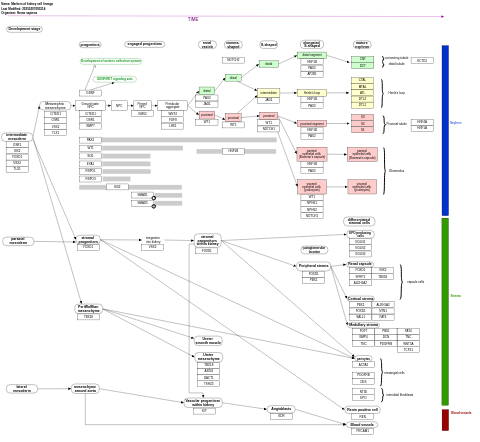
- Markers of kidney cell lineage - WP5236 (Homo sapiens)
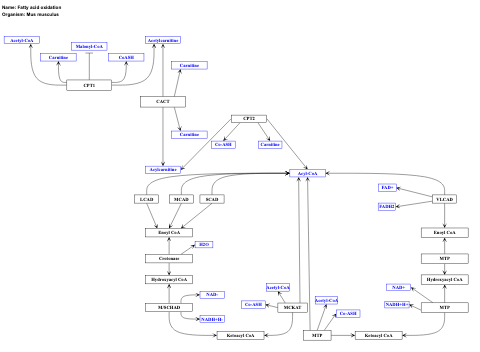
- Fatty acid oxidation - WP2318 (Mus musculus)
- Beta-alanine biosynthesis - WP2207 (Oryza sativa)
- Starch and cellulose biosynthesis - WP257 (Saccharomyces cerevisiae)
- Ureide biosynthesis - WP617 (Oryza sativa)
- Complement activation - WP545 (Homo sapiens)
- Sulfur degradation - WP440 (Saccharomyces cerevisiae)
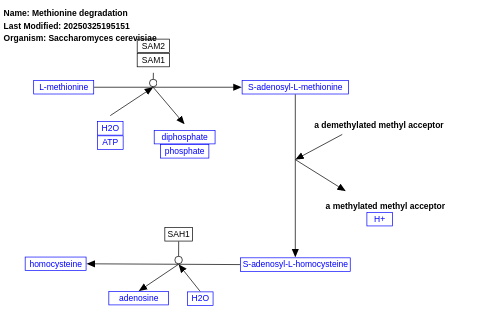
- Methionine degradation - WP46 (Saccharomyces cerevisiae)
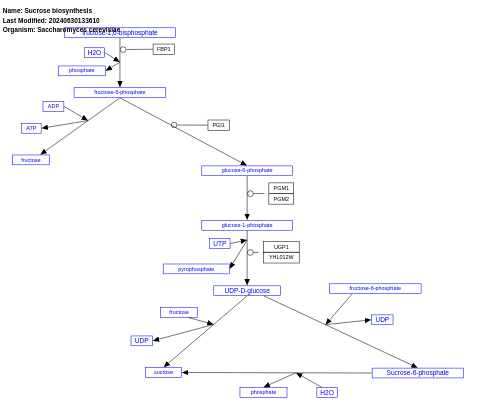
- Sucrose biosynthesis - WP14 (Saccharomyces cerevisiae)
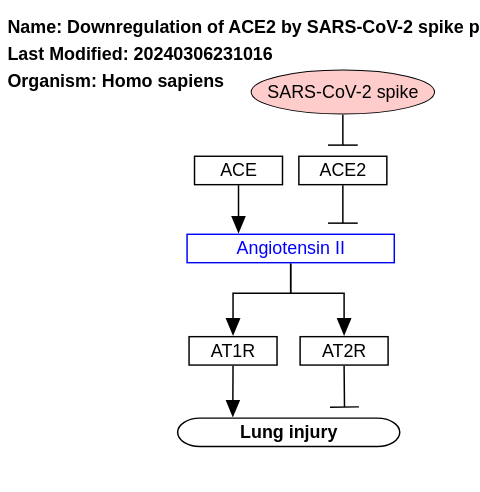
- Downregulation of ACE2 by SARS-CoV-2 spike protein - WP4799 (Homo sapiens)
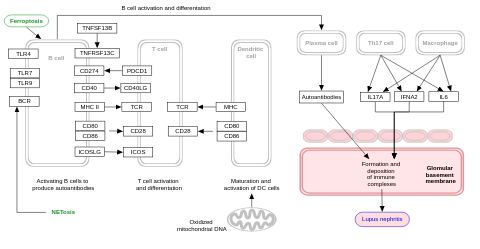
- Lupus pathogenesis - WP5559 (Homo sapiens)
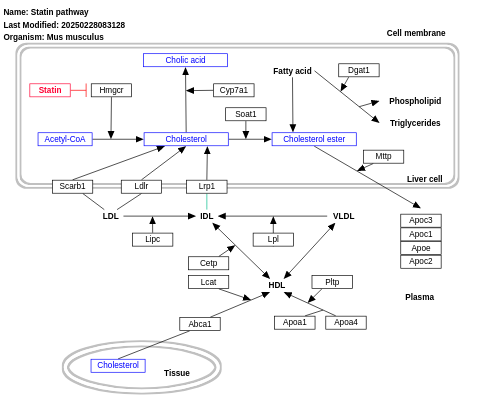
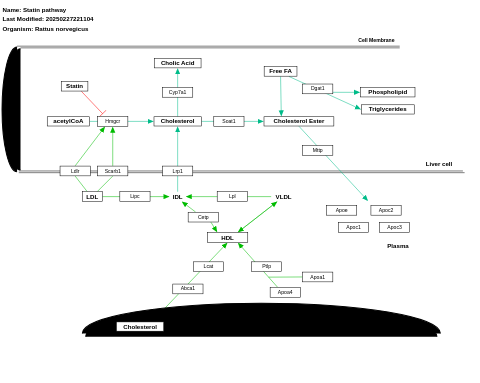
- Statin pathway - WP1 (Mus musculus)
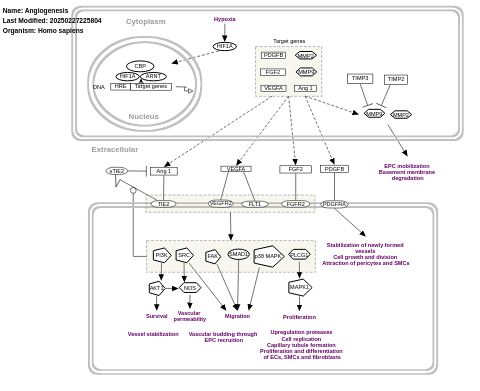
- Angiogenesis - WP1539 (Homo sapiens)
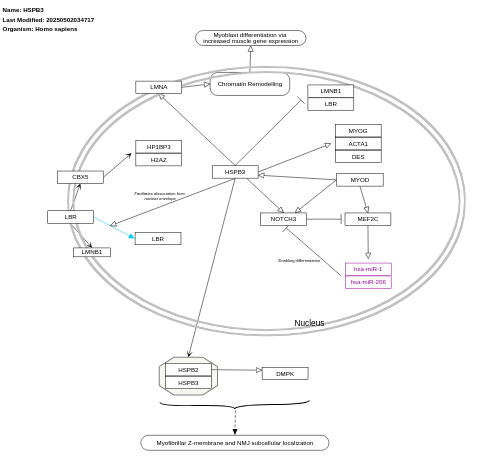
- HSPB3 in Charcot-Marie-Tooth disease - WP5524 (Homo sapiens)
- Nonalcoholic fatty liver disease - WP4396 (Homo sapiens)
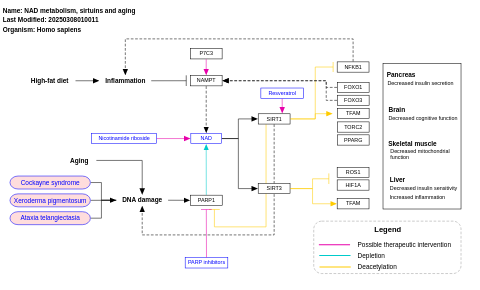
- NAD metabolism, sirtuins and aging - WP3630 (Homo sapiens)
- Prostaglandin synthesis and regulation - WP98 (Homo sapiens)
- Senescence and autophagy in cancer - WP615 (Homo sapiens)
- IL23 inhibitors in inflammatory bowel disease - WP5516 (Homo sapiens)
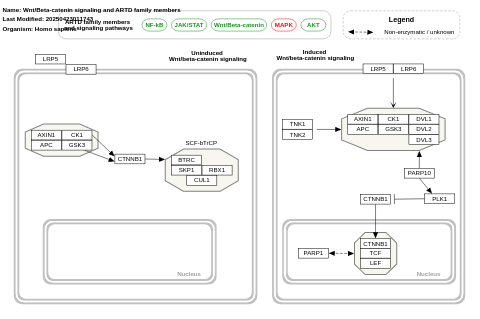
- MAPK signaling and ARTD family members - WP5530 (Homo sapiens)
- Wnt/Beta-catenin signaling and ARTD family members - WP5529 (Homo sapiens)
- Androgen receptor network in prostate cancer - WP2263 (Homo sapiens)
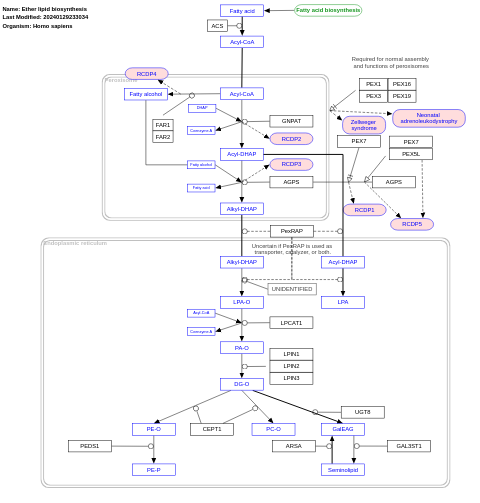
- Ether lipid biosynthesis - WP5275 (Homo sapiens)
- Primary focal segmental glomerulosclerosis (FSGS) - WP2572 (Homo sapiens)
- GABA receptor signaling - WP4159 (Homo sapiens)
- ATM signaling - WP2516 (Homo sapiens)
- Wnt signaling, NetPath - WP363 (Homo sapiens)
- Cholesterol biosynthesis pathway - WP197 (Homo sapiens)
- Vitamin A and carotenoid metabolism - WP716 (Homo sapiens)
- Pluripotent stem cell differentiation pathway - WP2848 (Homo sapiens)
- Parkinson's disease pathway - WP2371 (Homo sapiens)
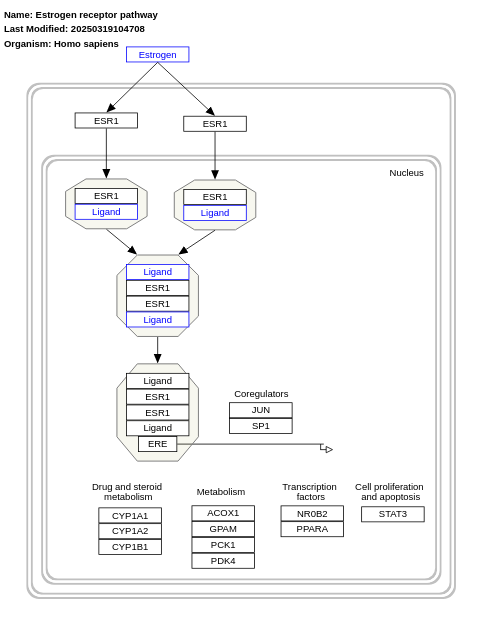
- Estrogen receptor pathway - WP2881 (Homo sapiens)
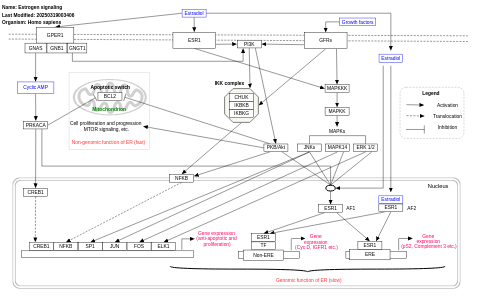
- Estrogen signaling - WP712 (Homo sapiens)
- Metabolic pathways of fibroblasts - WP5312 (Homo sapiens)
- DNA replication - WP466 (Homo sapiens)
- One-carbon metabolism - WP435 (Mus musculus)
- Complement system - WP2806 (Homo sapiens)
- One-carbon metabolism - WP241 (Homo sapiens)
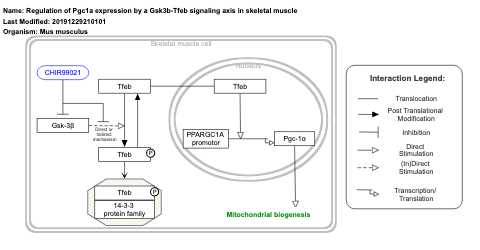
- Regulation of Pgc1a expression by a Gsk3b-Tfeb signaling axis in skeletal muscle - WP4763 (Mus musculus)
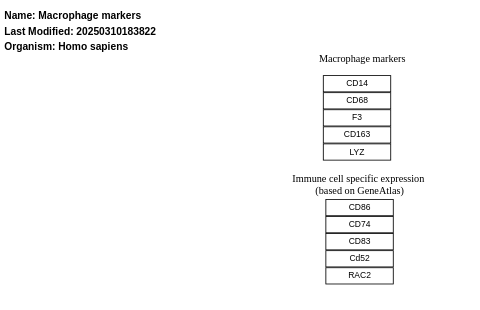
- Macrophage markers - WP4146 (Homo sapiens)
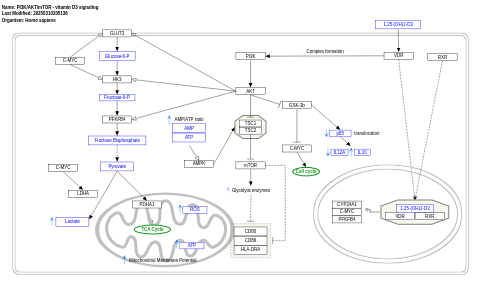
- PI3K/AKT/mTOR - vitamin D3 signaling - WP4141 (Homo sapiens)
- Angiopoietin-like protein 8 regulatory pathway - WP3915 (Homo sapiens)
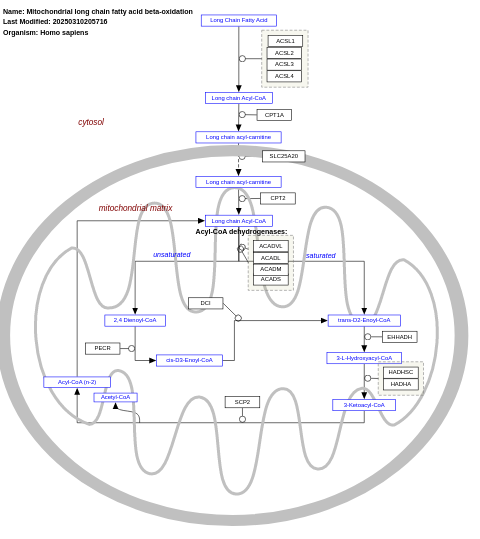
- Mitochondrial long chain fatty acid beta-oxidation - WP368 (Homo sapiens)
- Interactions between LOXL4 and oxidative stress pathway - WP3670 (Homo sapiens)
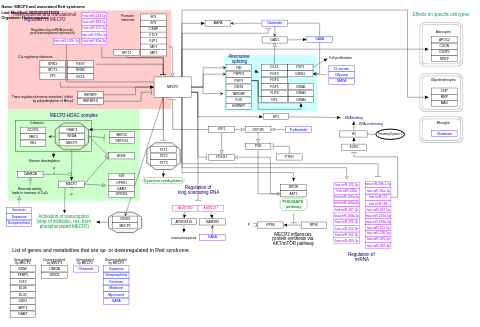
- MECP2 and associated Rett syndrome - WP3584 (Homo sapiens)
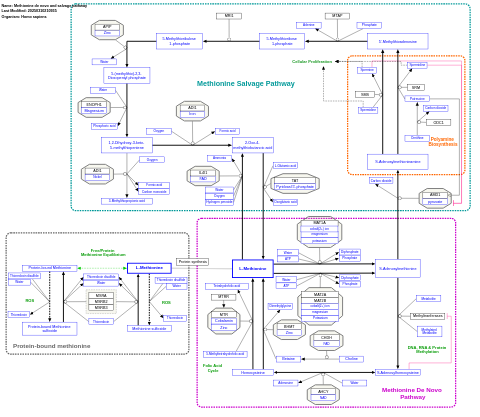
- Methionine de novo and salvage pathway - WP3580 (Homo sapiens)
- Fatty acid biosynthesis - WP357 (Homo sapiens)
- Melatonin metabolism and effects - WP3298 (Homo sapiens)
- Corticotropin-releasing hormone - WP3265 (Bos taurus)
- Oncostatin M signaling - WP2374 (Homo sapiens)
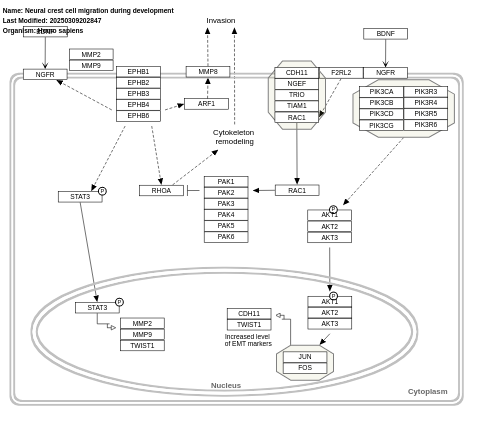
- Neural crest cell migration during development - WP4564 (Homo sapiens)
- Parkin-ubiquitin proteasomal system pathway - WP2359 (Homo sapiens)
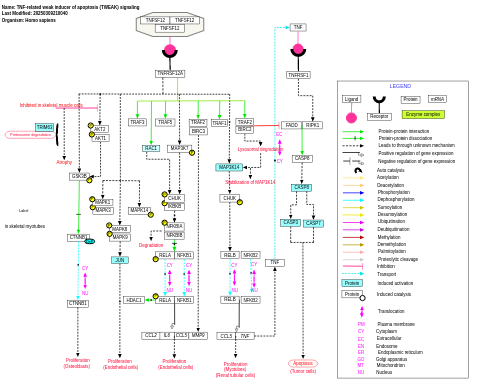
- TNF-related weak inducer of apoptosis (TWEAK) signaling - WP2036 (Homo sapiens)
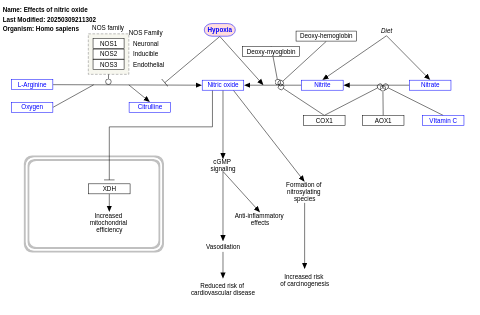
- Effects of nitric oxide - WP1995 (Homo sapiens)
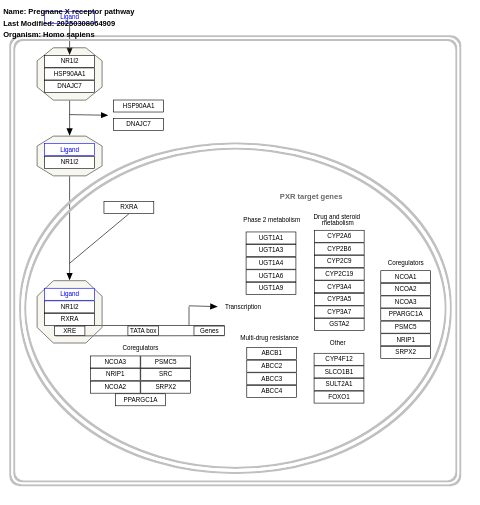
- Pregnane X receptor pathway - WP2876 (Homo sapiens)
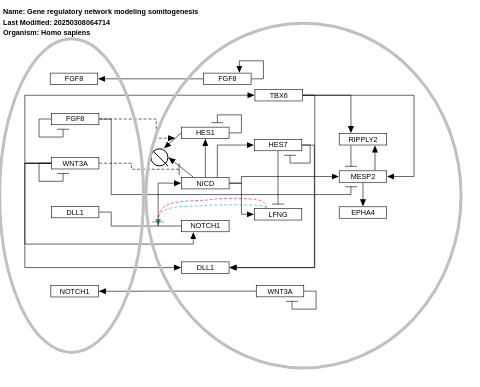
- Gene regulatory network modeling somitogenesis - WP2854 (Homo sapiens)
- Endoderm differentiation - WP2853 (Homo sapiens)
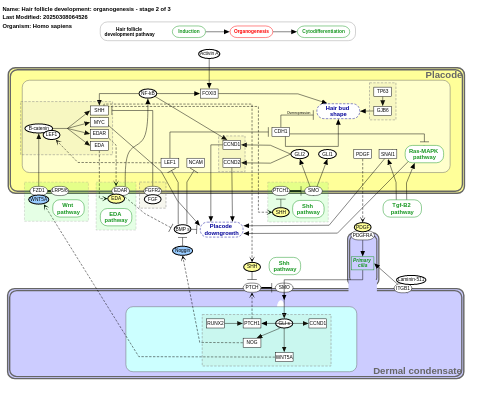
- Hair follicle development: organogenesis - stage 2 of 3 - WP2839 (Homo sapiens)
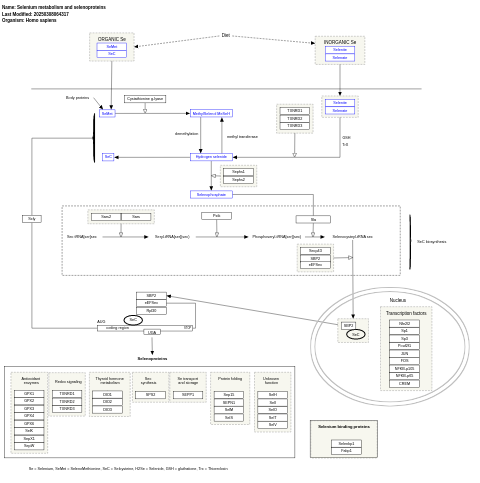
- Selenium metabolism and selenoproteins - WP28 (Homo sapiens)
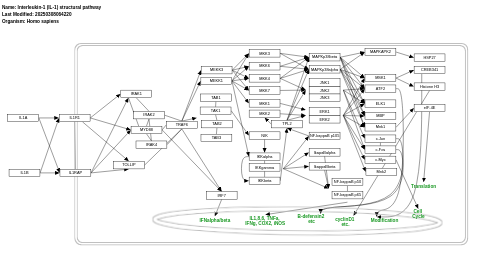
- Interleukin-1 (IL-1) structural pathway - WP2637 (Homo sapiens)
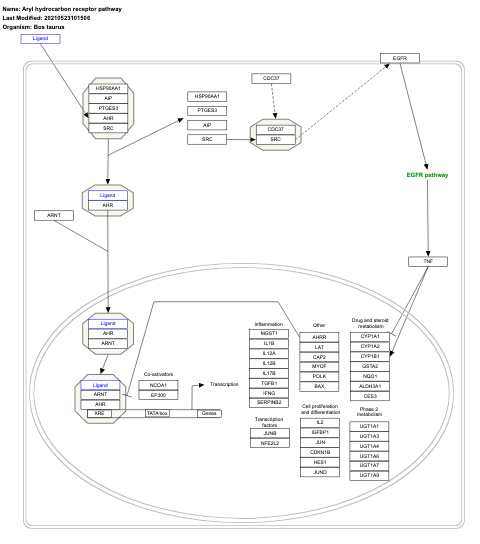
- Aryl hydrocarbon receptor pathway - WP2586 (Homo sapiens)
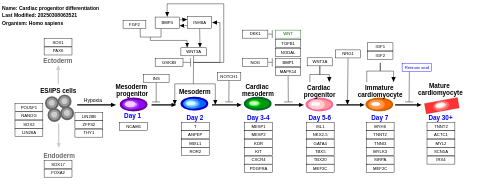
- Cardiac progenitor differentiation - WP2406 (Homo sapiens)
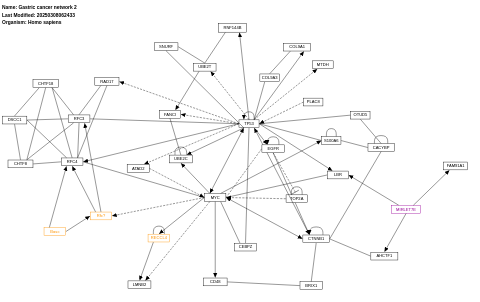
- Gastric cancer network 2 - WP2363 (Homo sapiens)
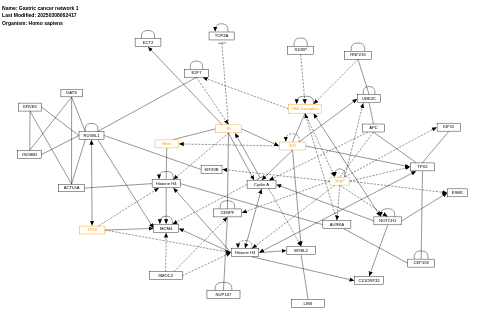
- Gastric cancer network 1 - WP2361 (Homo sapiens)
- Adipogenesis - WP236 (Homo sapiens)
- Corticotropin-releasing hormone signaling - WP2355 (Homo sapiens)
- miRNA biogenesis - WP2338 (Homo sapiens)
- AGE/RAGE pathway - WP2324 (Homo sapiens)
- Chemokine signaling pathway - WP2292 (Mus musculus)
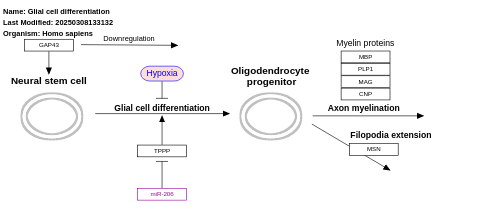
- Glial cell differentiation - WP2276 (Homo sapiens)
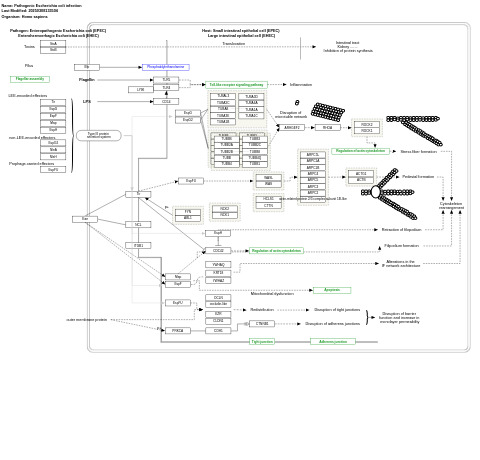
- Pathogenic Escherichia coli infection - WP2272 (Homo sapiens)
- Glioblastoma signaling - WP2261 (Homo sapiens)
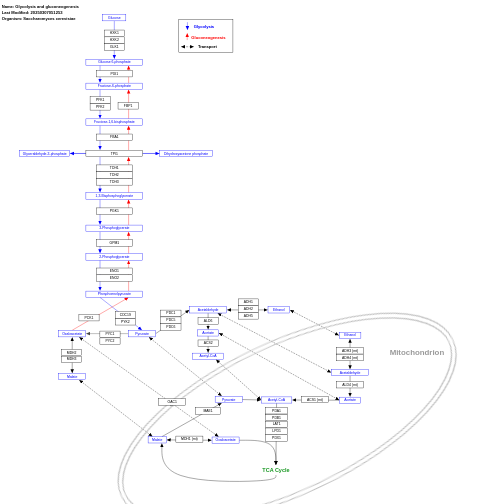
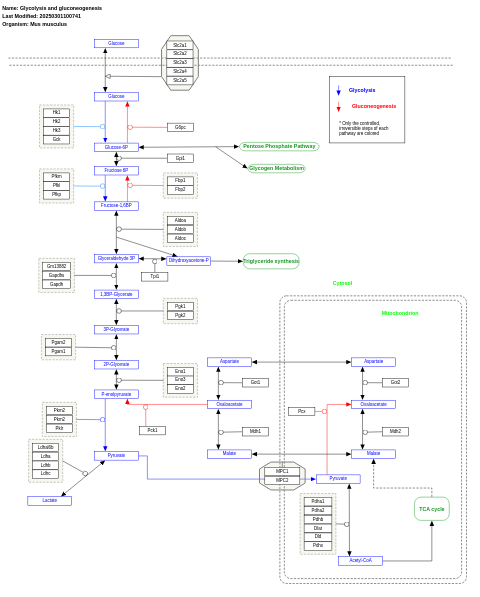
- Glycolysis and gluconeogenesis - WP515 (Saccharomyces cerevisiae)
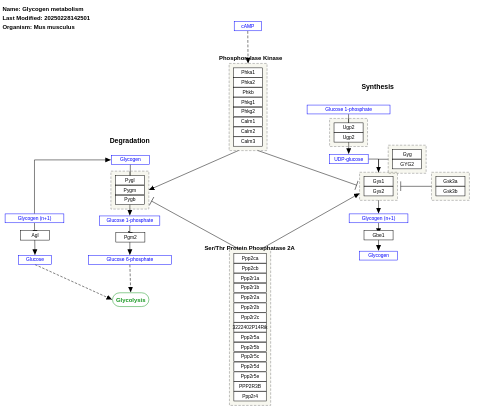
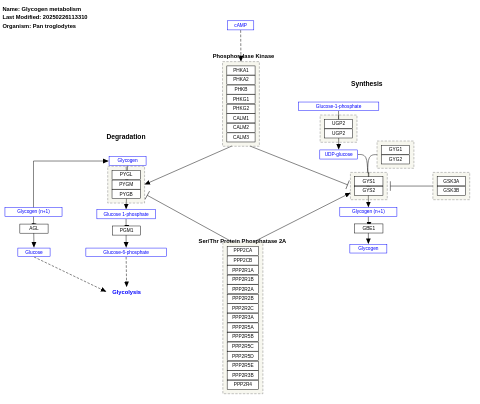
- Glycogen synthesis and degradation - WP500 (Homo sapiens)
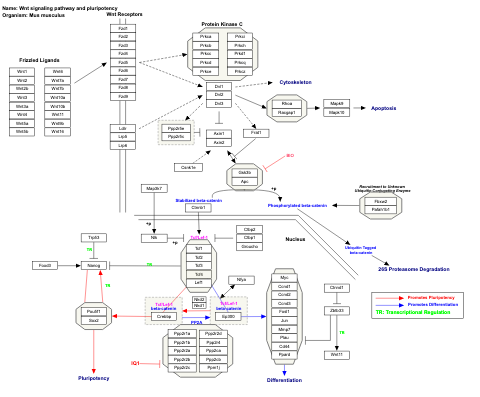
- Wnt signaling and pluripotency - WP399 (Homo sapiens)
- NAD+ metabolism - WP3644 (Homo sapiens)
- Parkinson's disease - WP3638 (Mus musculus)
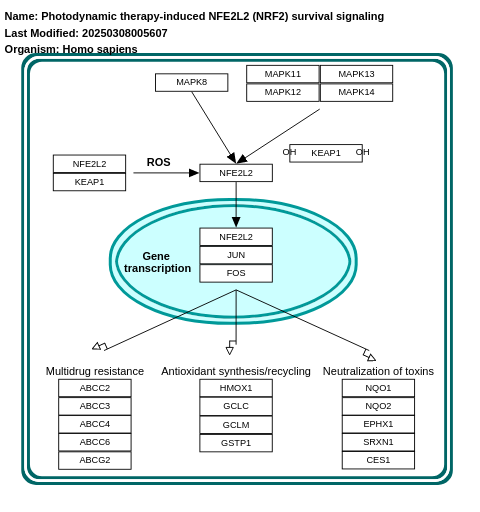
- Photodynamic therapy-induced NFE2L2 (NRF2) survival signaling - WP3612 (Homo sapiens)
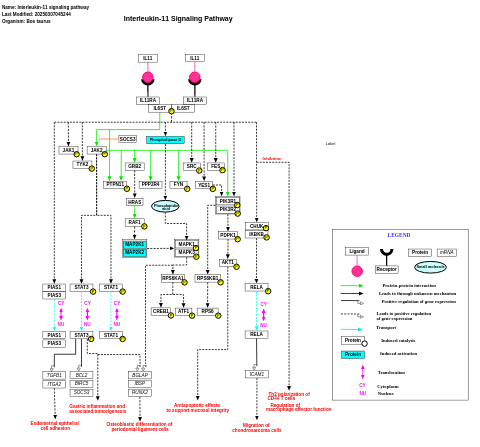
- Interleukin-11 signaling pathway - WP3159 (Bos taurus)
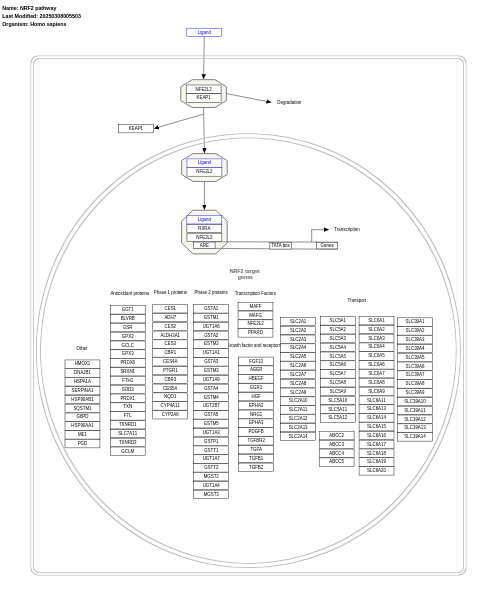
- NRF2 pathway - WP2884 (Homo sapiens)
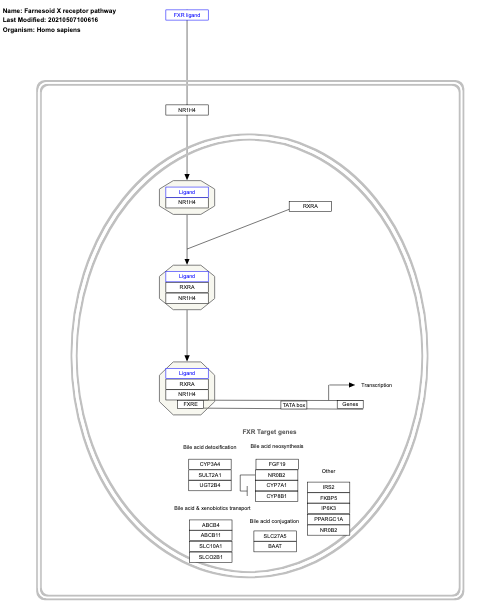
- Farnesoid X receptor pathway - WP2879 (Homo sapiens)
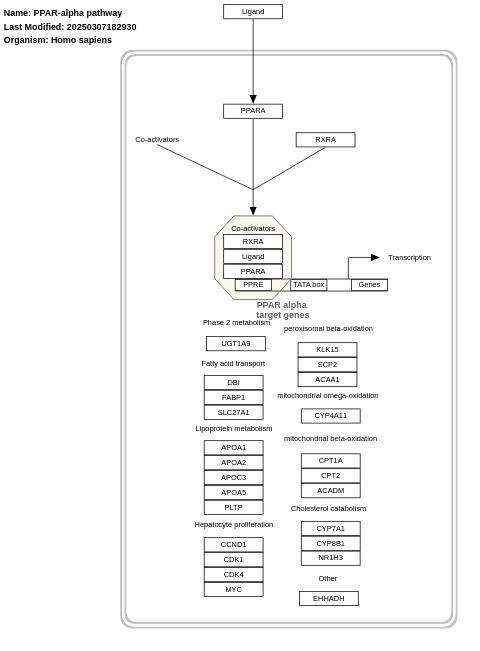
- PPAR-alpha pathway - WP2878 (Homo sapiens)
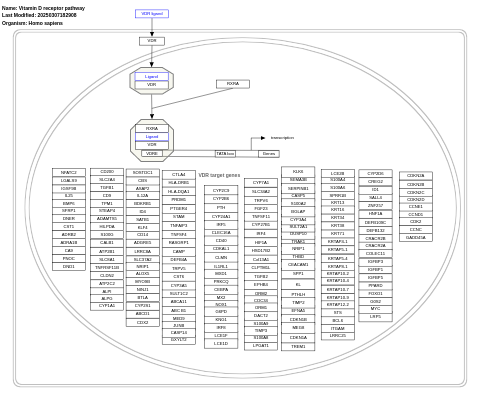
- Vitamin D receptor pathway - WP2877 (Homo sapiens)
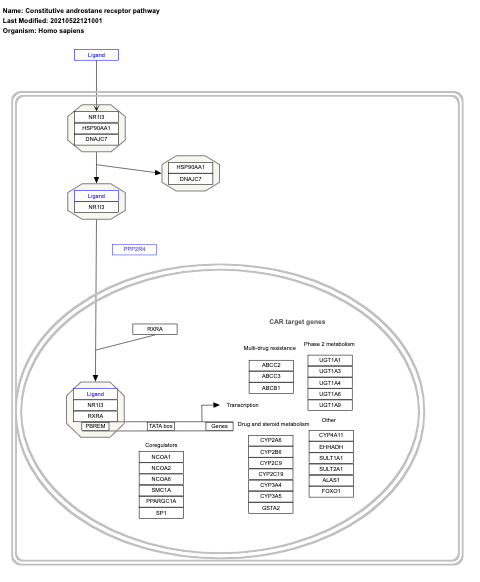
- Constitutive androstane receptor pathway - WP2875 (Homo sapiens)
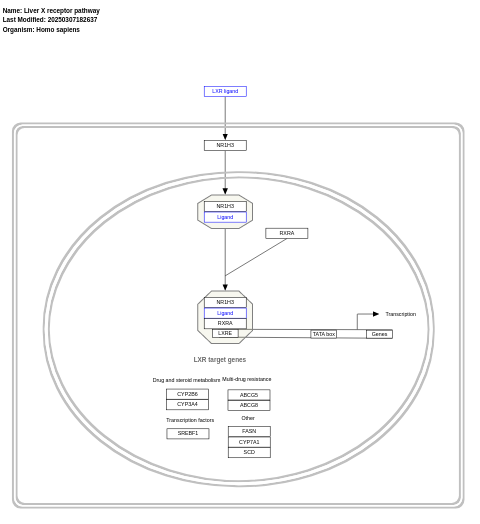
- Liver X receptor pathway - WP2874 (Homo sapiens)
- Aryl hydrocarbon receptor pathway - WP2873 (Homo sapiens)
- Thymic stromal lymphopoietin (TSLP) signaling - WP2203 (Homo sapiens)
- IL9 signaling - WP22 (Homo sapiens)
- Arrhythmogenic right ventricular cardiomyopathy - WP2118 (Homo sapiens)
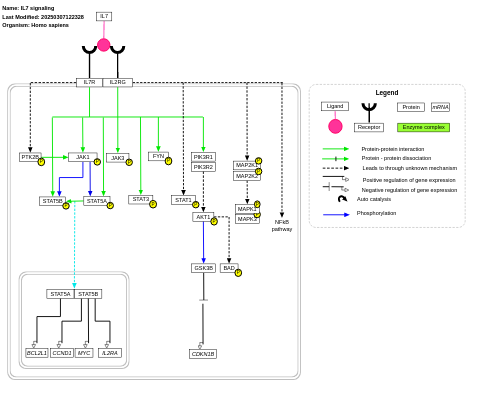
- IL7 signaling - WP205 (Homo sapiens)
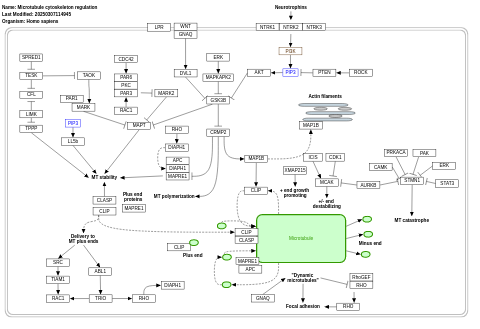
- Microtubule cytoskeleton regulation - WP2038 (Homo sapiens)
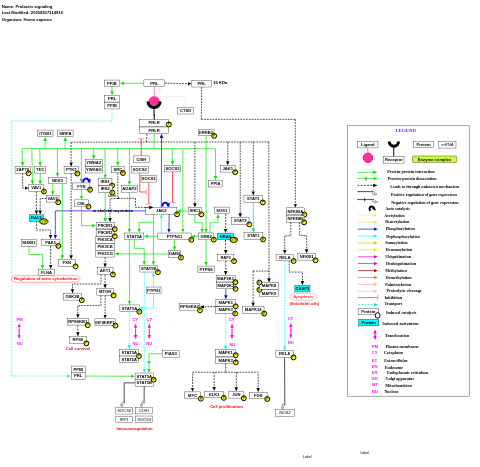
- Prolactin signaling - WP2037 (Homo sapiens)
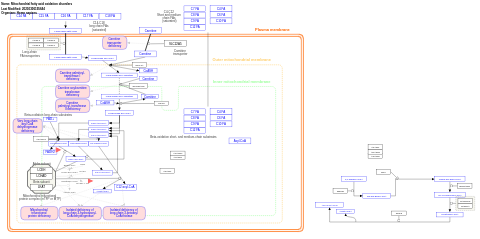
- Mitochondrial fatty acid oxidation disorders - WP5123 (Homo sapiens)
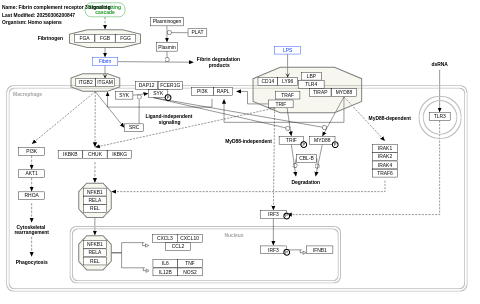
- Fibrin complement receptor 3 signaling - WP4136 (Homo sapiens)
- Liver steatosis adverse outcome pathway - WP4010 (Homo sapiens)
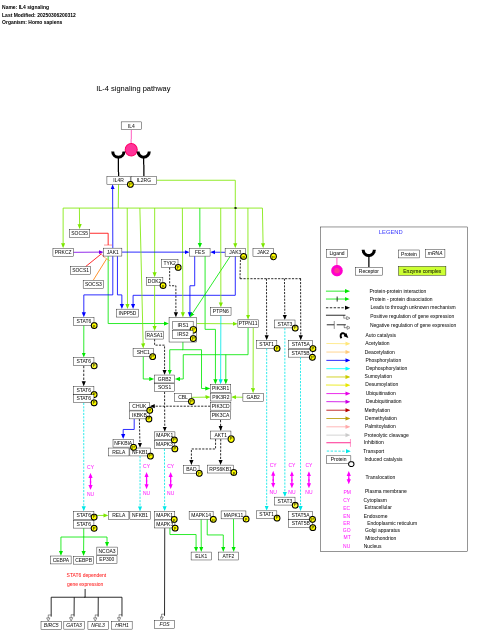
- IL4 signaling - WP395 (Homo sapiens)
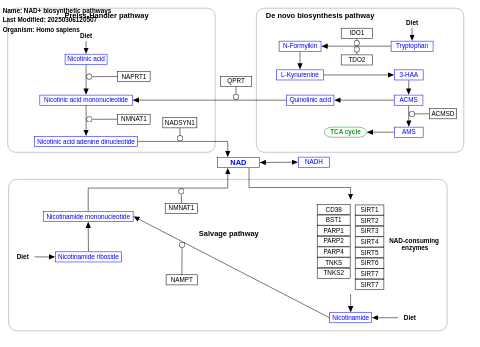
- NAD+ biosynthetic pathways - WP3645 (Homo sapiens)
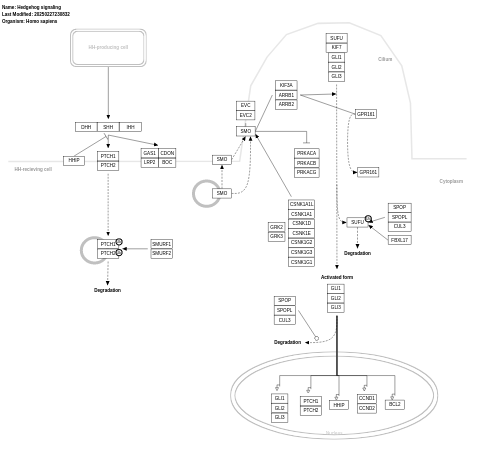
- Hedgehog signaling - WP47 (Homo sapiens)
- Type 2 papillary renal cell carcinoma - WP4241 (Homo sapiens)
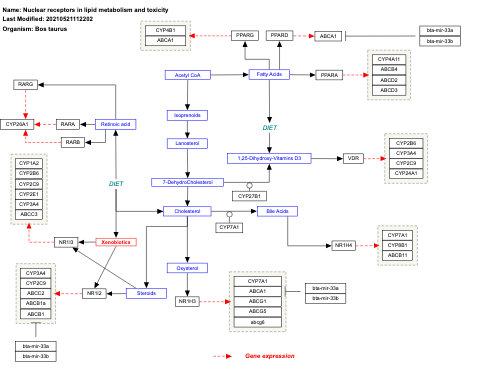
- Nuclear receptors in lipid metabolism and toxicity - WP299 (Homo sapiens)
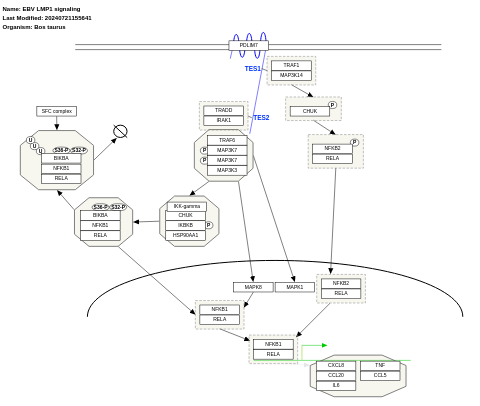
- Ebstein-Barr virus LMP1 signaling - WP262 (Homo sapiens)
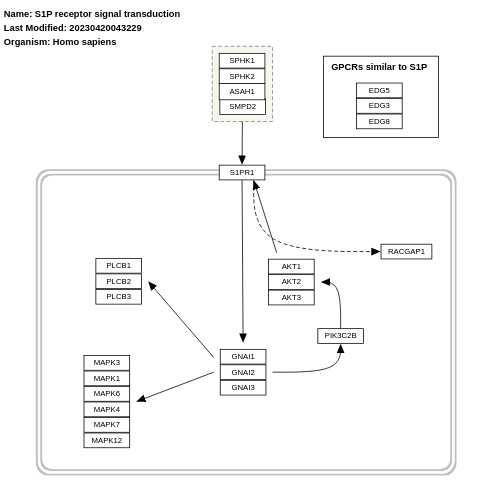
- S1P receptor signal transduction - WP26 (Homo sapiens)
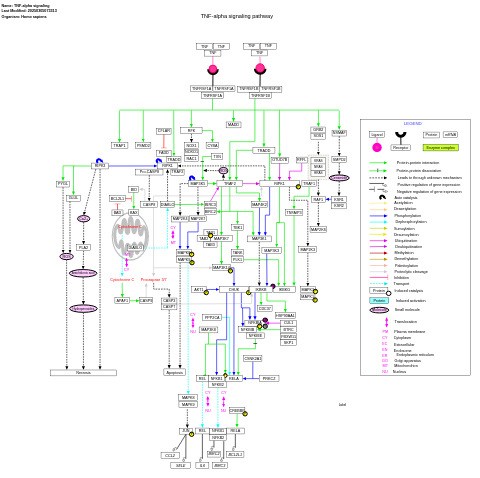
- TNF-alpha signaling - WP231 (Homo sapiens)
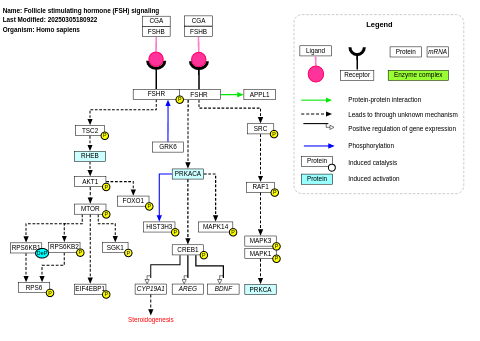
- Follicle stimulating hormone (FSH) signaling - WP2035 (Homo sapiens)
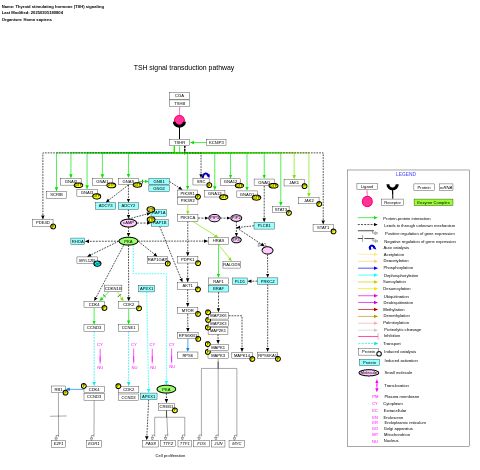
- Thyroid stimulating hormone (TSH) signaling - WP2032 (Homo sapiens)
- Cell differentiation - index - WP2029 (Homo sapiens)
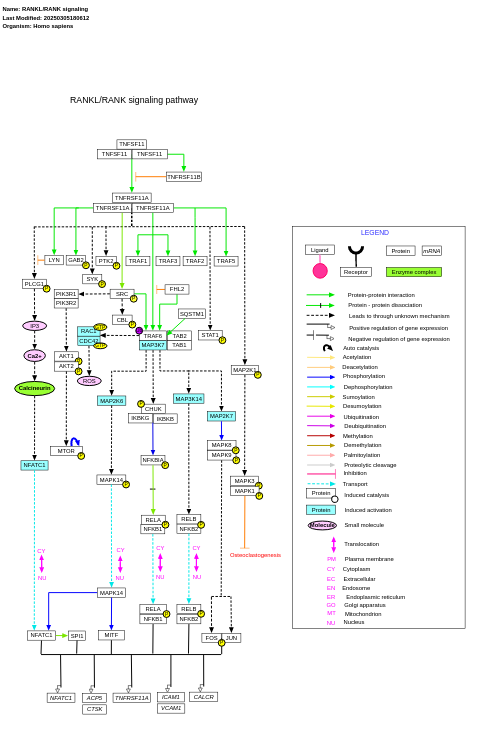
- RANKL/RANK signaling - WP2018 (Homo sapiens)
- Integrated breast cancer pathway - WP1984 (Homo sapiens)
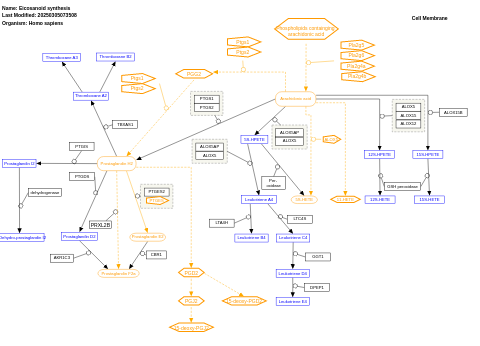
- Eicosanoid synthesis - WP167 (Homo sapiens)
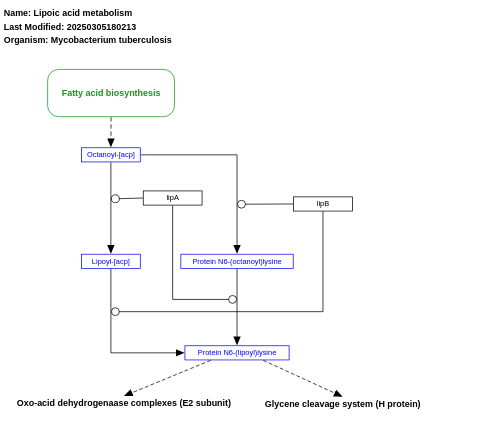
- Lipoic acid metabolism - WP1667 (Mycobacterium tuberculosis)
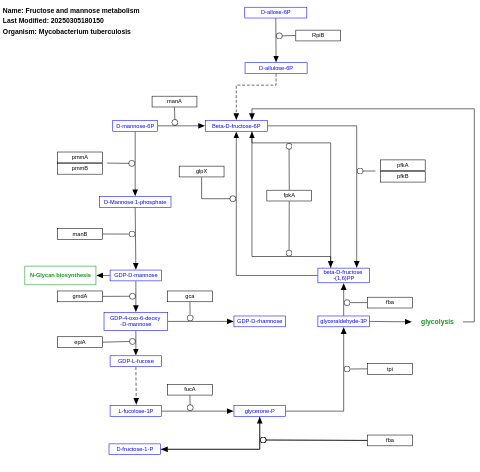
- Fructose and mannose metabolism - WP1652 (Mycobacterium tuberculosis)
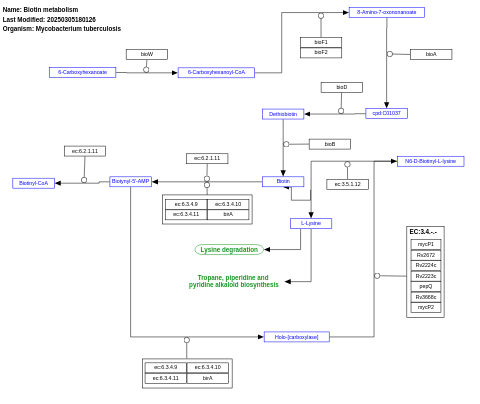
- Biotin metabolism - WP1631 (Mycobacterium tuberculosis)
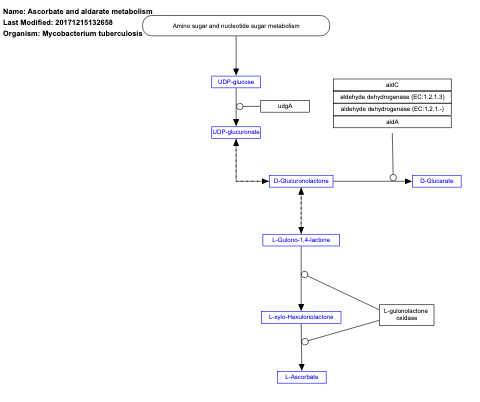
- Ascorbate and aldarate metabolism - WP1622 (Mycobacterium tuberculosis)
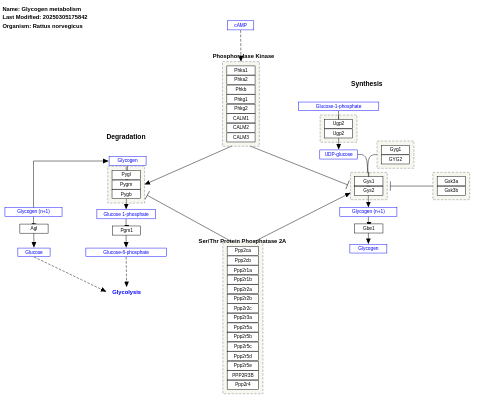
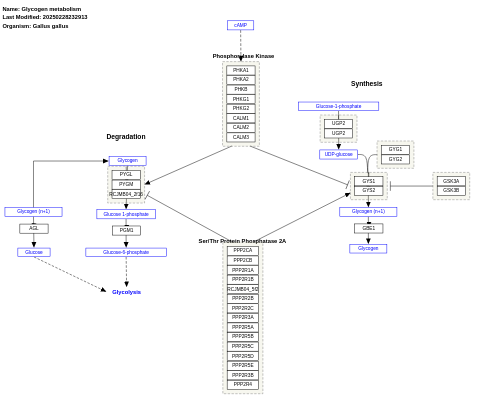
- Glycogen metabolism - WP160 (Rattus norvegicus)
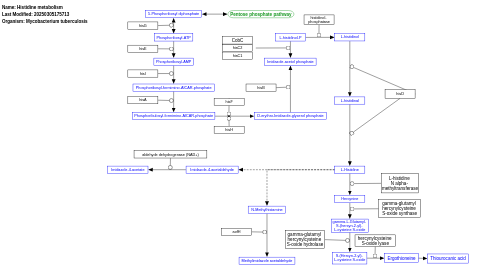
- Histidine metabolism - WP1581 (Mycobacterium tuberculosis)
- MicroRNAs in cardiomyocyte hypertrophy - WP1544 (Homo sapiens)
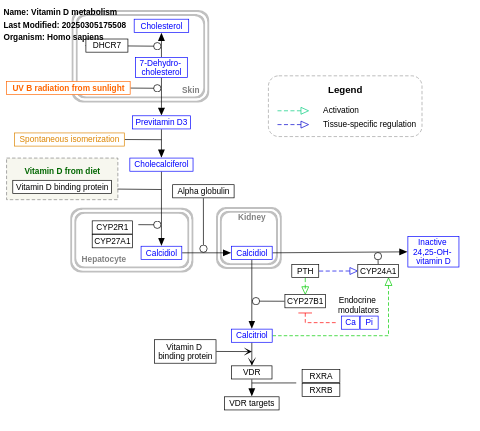
- Vitamin D metabolism - WP1531 (Homo sapiens)
- Glucuronidation - WP698 (Homo sapiens)
- Estrogen metabolism - WP697 (Homo sapiens)
- Benzo(a)pyrene metabolism - WP696 (Homo sapiens)
- Sulfation biotransformation reaction - WP692 (Homo sapiens)
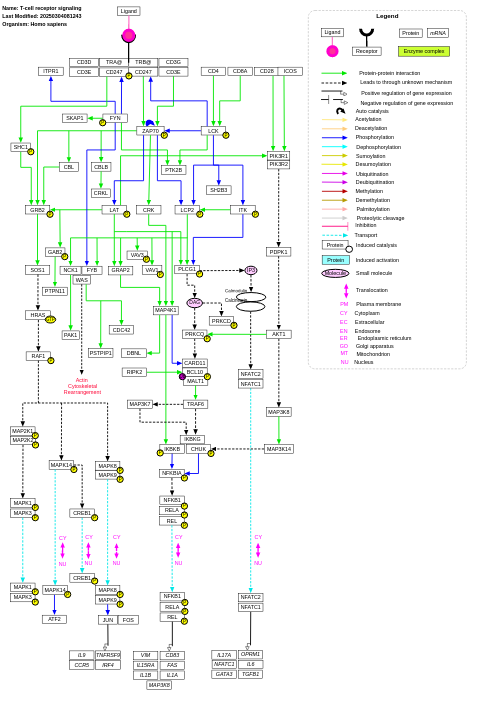
- T-cell receptor signaling - WP69 (Homo sapiens)
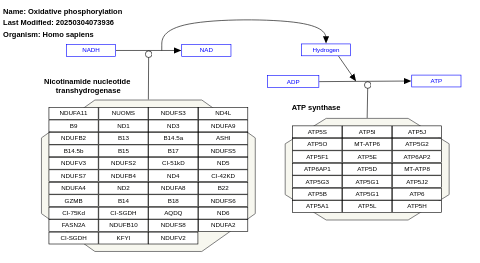
- Oxidative phosphorylation - WP623 (Homo sapiens)
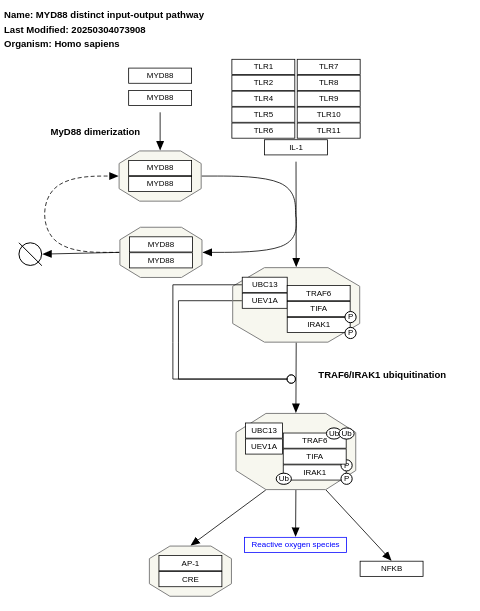
- MYD88 distinct input-output pathway - WP3877 (Homo sapiens)
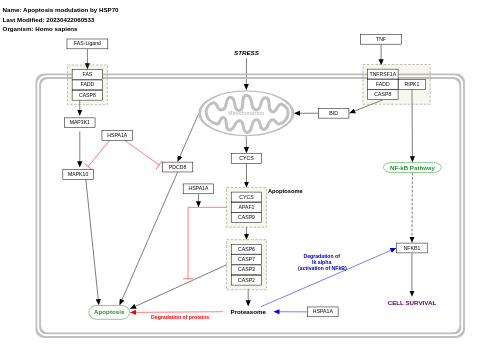
- Apoptosis modulation by HSP70 - WP384 (Homo sapiens)
- EGF/EGFR signaling pathway - WP978 (Bos taurus)
- Androgen receptor signaling pathway - WP897 (Pan troglodytes)
- Glycogen metabolism - WP835 (Gallus gallus)
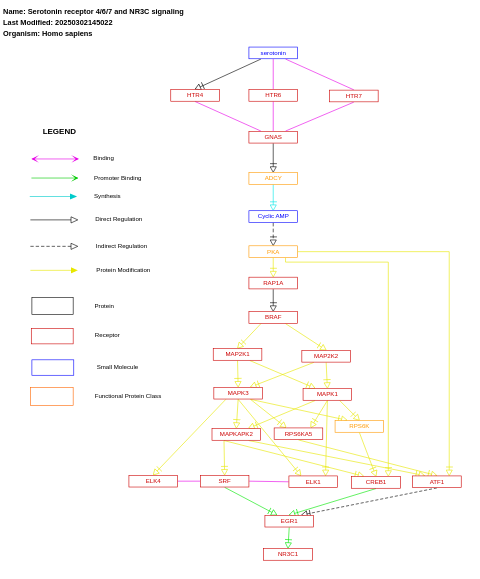
- Serotonin receptor 4/6/7 and NR3C signaling - WP734 (Homo sapiens)
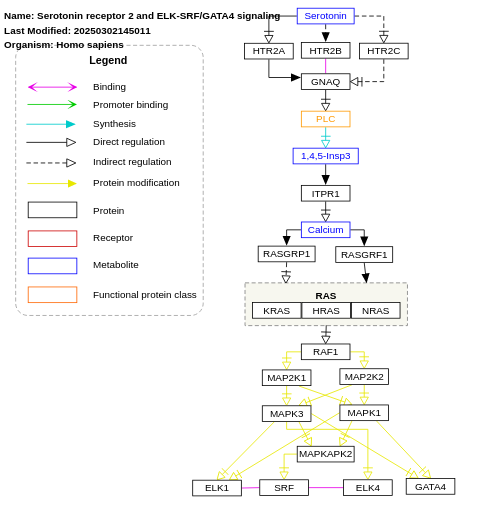
- Serotonin receptor 2 and ELK-SRF/GATA4 signaling - WP732 (Homo sapiens)
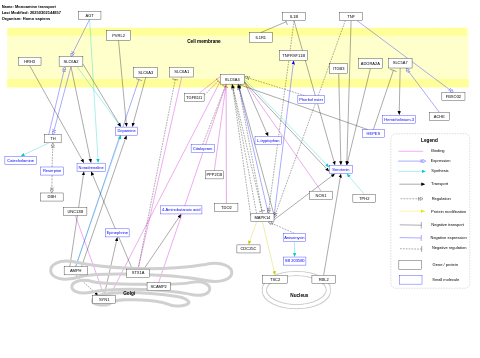
- Monoamine transport - WP727 (Homo sapiens)
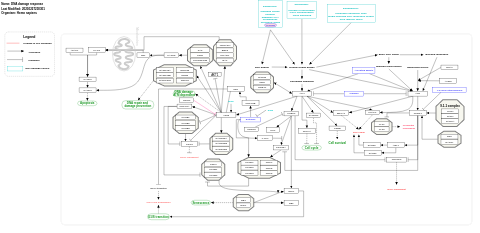
- DNA damage response - WP707 (Homo sapiens)
- Sudden infant death syndrome (SIDS) susceptibility pathways - WP706 (Homo sapiens)
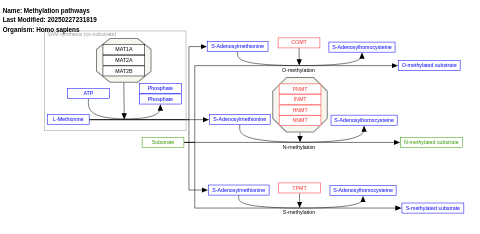
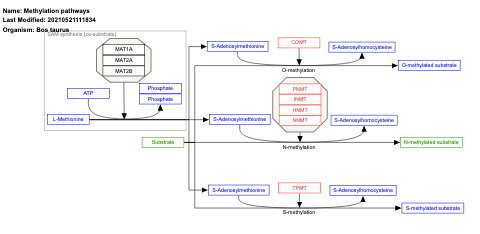
- Methylation pathways - WP704 (Homo sapiens)
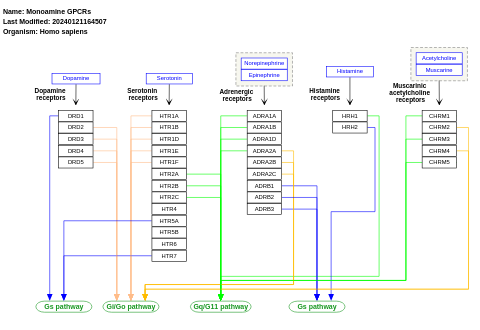
- Monoamine GPCRs - WP58 (Homo sapiens)
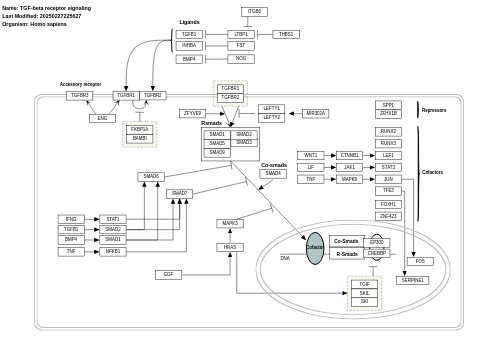
- TGF-beta receptor signaling - WP560 (Homo sapiens)
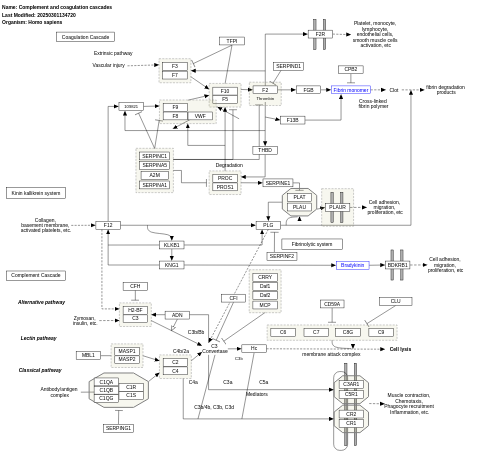
- Complement and coagulation cascades - WP558 (Homo sapiens)
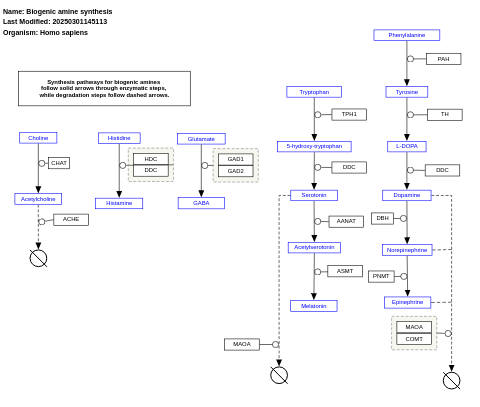
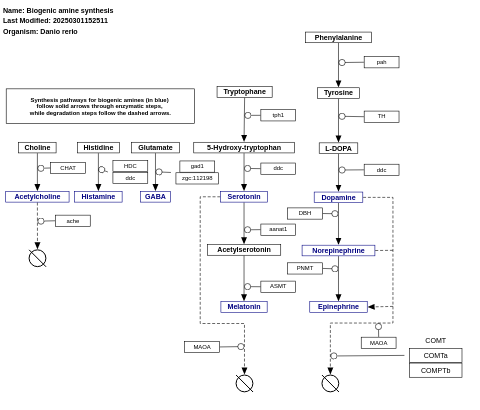
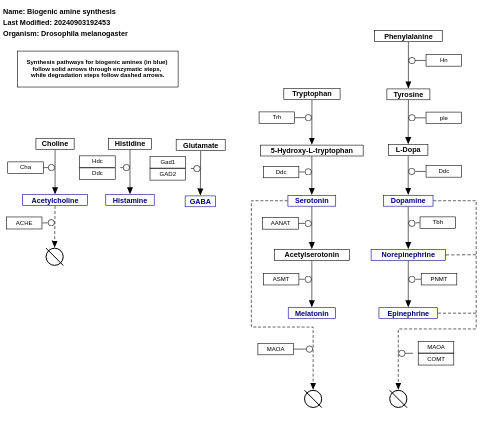
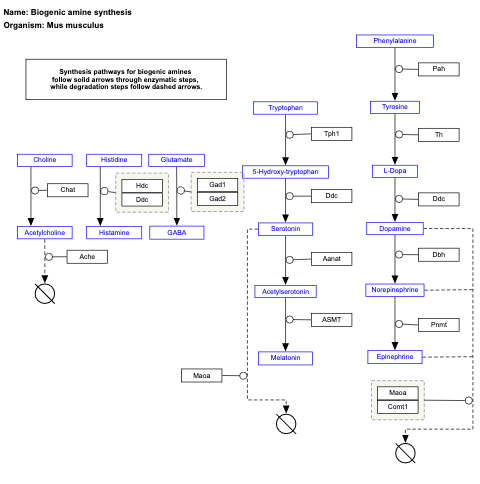
- Biogenic amine synthesis - WP550 (Homo sapiens)
- Complement and coagulation cascades - WP547 (Rattus norvegicus)
- Glycolysis and gluconeogenesis - WP534 (Homo sapiens)
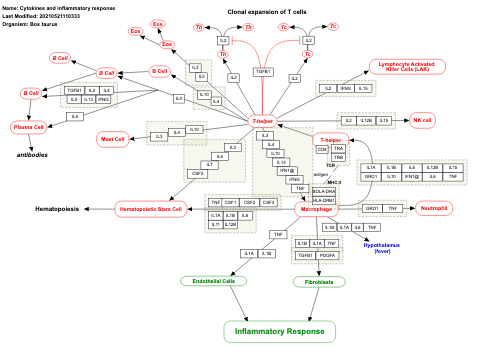
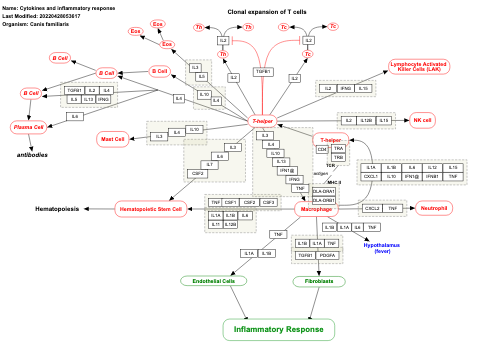
- Cytokines and inflammatory response - WP530 (Homo sapiens)
- Glucose metabolism in triple-negative breast cancer cells - WP5211 (Homo sapiens)
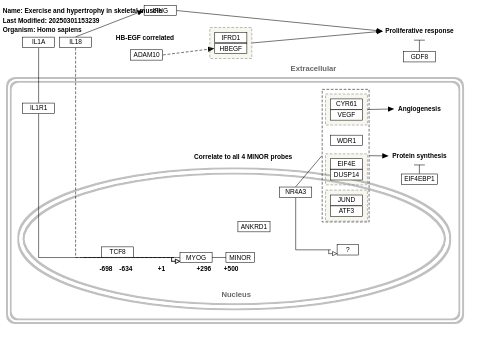
- Exercise and hypertrophy in skeletal muscle - WP516 (Homo sapiens)
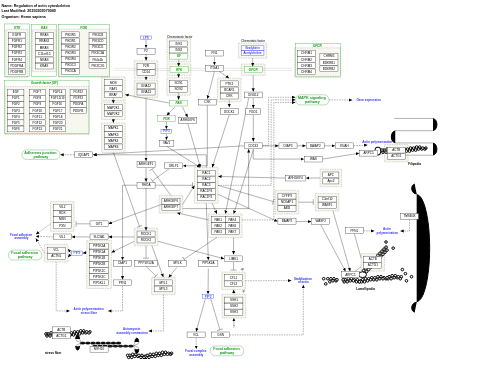
- Regulation of actin cytoskeleton - WP51 (Homo sapiens)
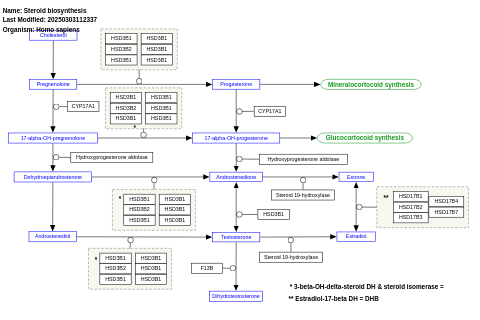
- Steroid biosynthesis - WP496 (Homo sapiens)
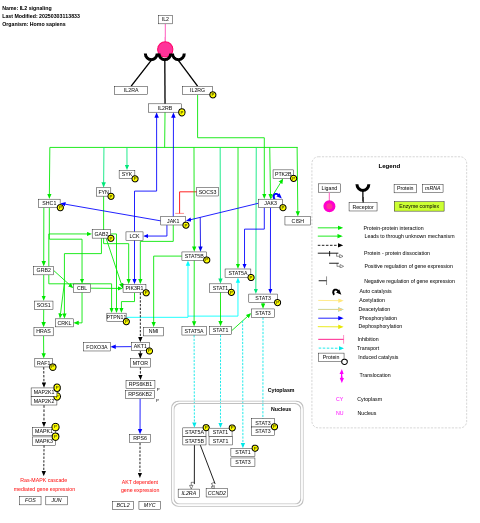
- IL2 signaling - WP49 (Homo sapiens)
- Ovarian infertility - WP34 (Homo sapiens)
- Peptide GPCRs - WP24 (Homo sapiens)
- B cell receptor signaling - WP23 (Homo sapiens)
- Fatty acid omega-oxidation - WP206 (Homo sapiens)
- miRNA regulation of DNA damage response - WP1530 (Homo sapiens)
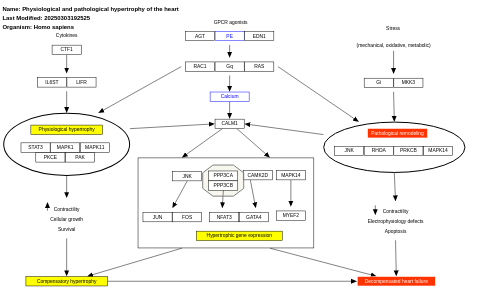
- Physiological and pathological hypertrophy of the heart - WP1528 (Homo sapiens)
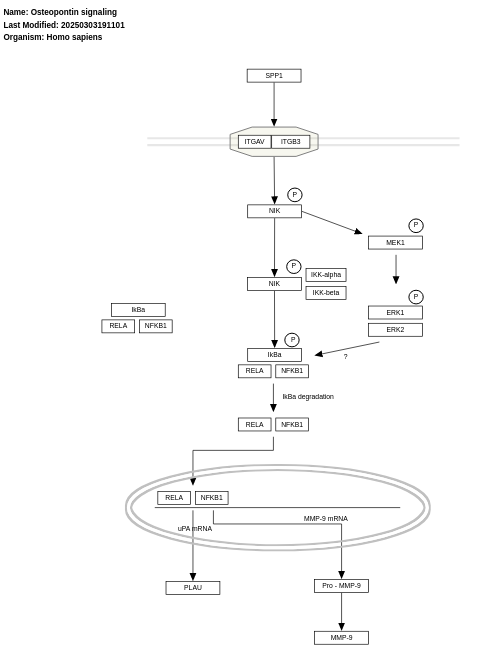
- Osteopontin signaling - WP1434 (Homo sapiens)
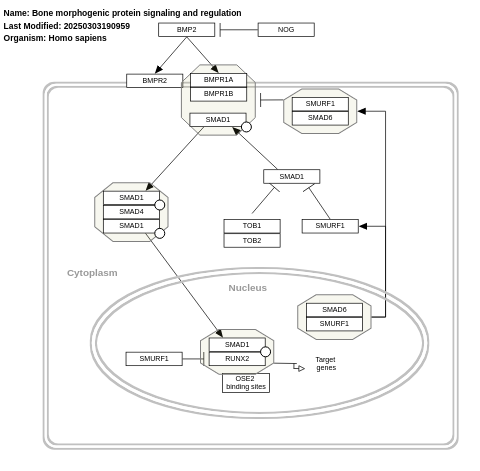
- Bone morphogenic protein signaling and regulation - WP1425 (Homo sapiens)
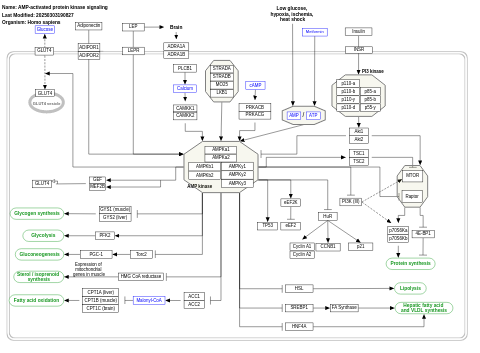
- AMP-activated protein kinase signaling - WP1403 (Homo sapiens)
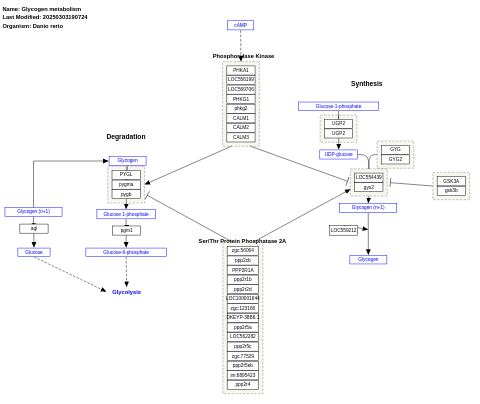
- Glycogen metabolism - WP1388 (Danio rerio)
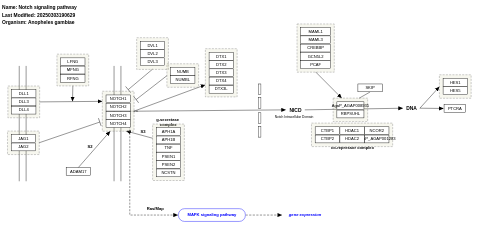
- Notch signaling pathway - WP1232 (Anopheles gambiae)
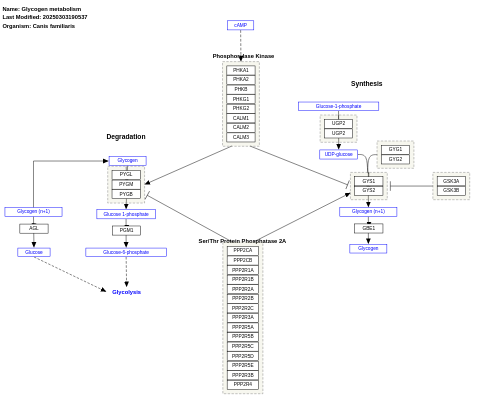
- Glycogen metabolism - WP1189 (Canis familiaris)
- Androgen receptor signaling pathway - WP1133 (Canis familiaris)
- Glycogen metabolism - WP1073 (Bos taurus)
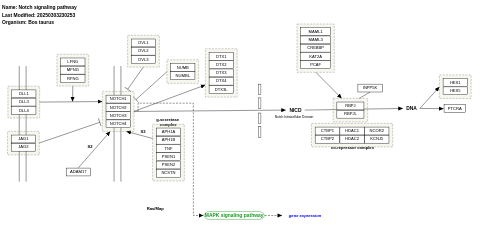
- Notch signaling pathway - WP1064 (Bos taurus)
- Alanine and aspartate metabolism - WP106 (Homo sapiens)
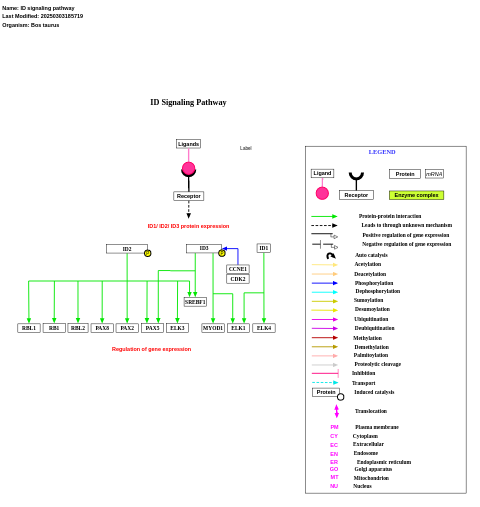
- ID signaling pathway - WP1052 (Bos taurus)
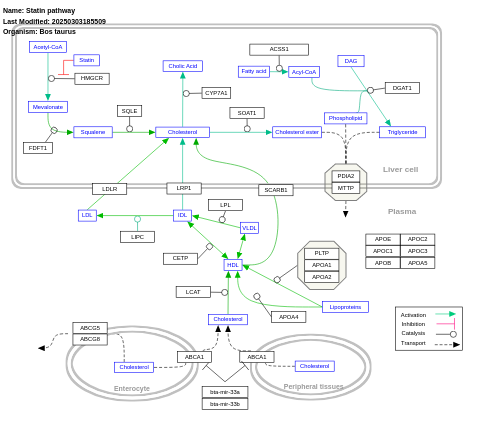
- Statin pathway - WP1041 (Bos taurus)
- Notch signaling pathway - WP1029 (Bos taurus)
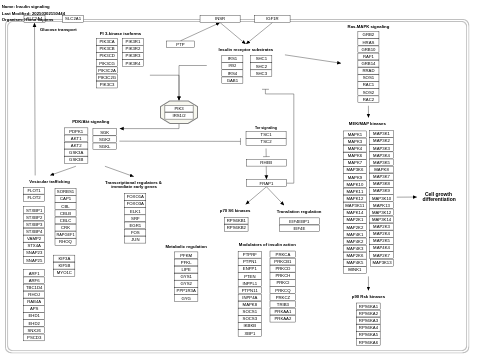
- Insulin signaling - WP481 (Homo sapiens)
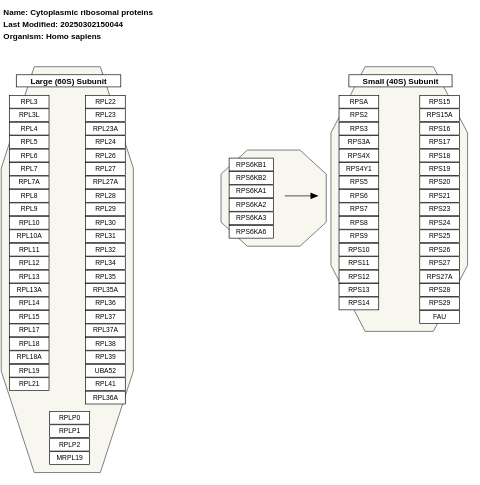
- Cytoplasmic ribosomal proteins - WP477 (Homo sapiens)
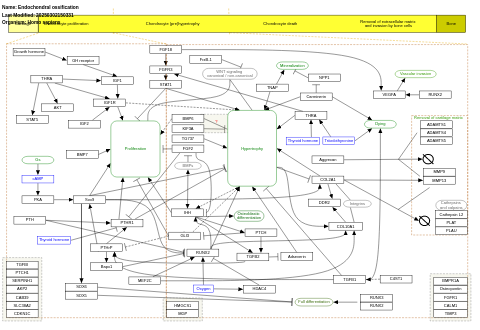
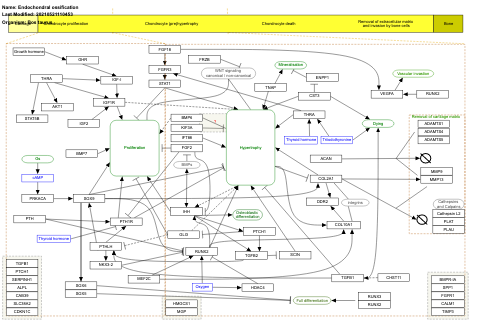
- Endochondral ossification - WP474 (Homo sapiens)
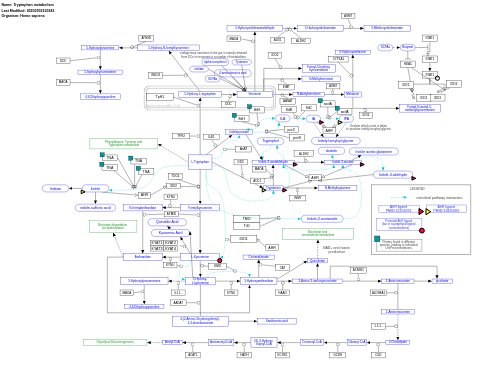
- Tryptophan metabolism - WP465 (Homo sapiens)
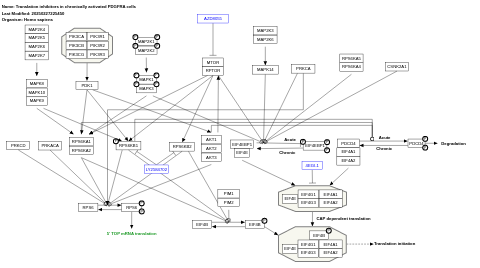
- Translation inhibitors in chronically activated PDGFRA cells - WP4566 (Homo sapiens)
- GPCRs, class A rhodopsin-like - WP455 (Homo sapiens)
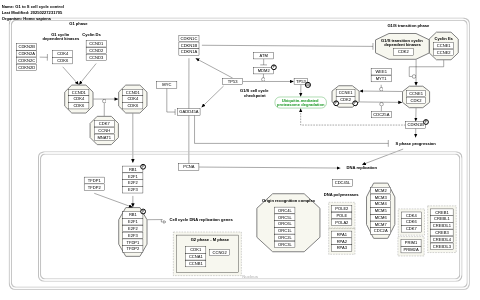
- G1 to S cell cycle control - WP45 (Homo sapiens)
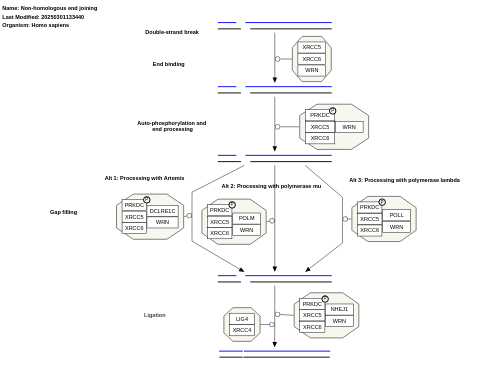
- Non-homologous end joining - WP438 (Homo sapiens)
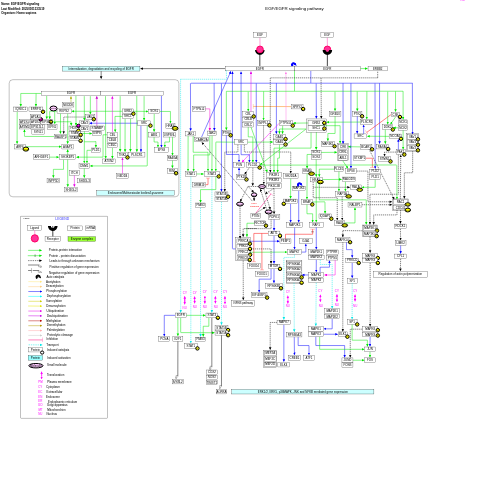
- EGF/EGFR signaling - WP437 (Homo sapiens)
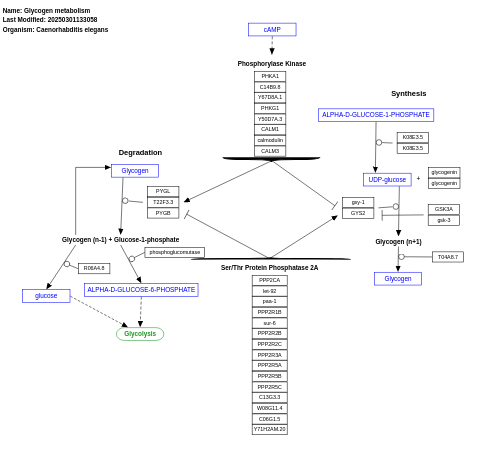
- Glycogen metabolism - WP436 (Caenorhabditis elegans)
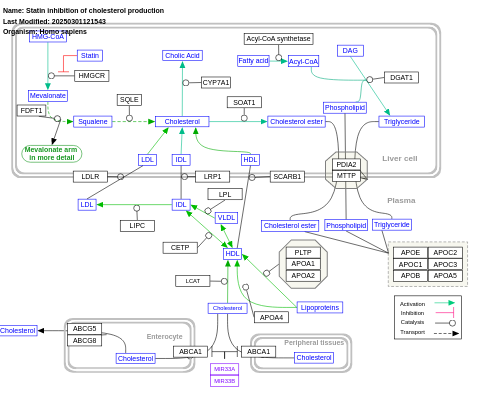
- Statin inhibition of cholesterol production - WP430 (Homo sapiens)
- Hedgehog signaling - WP4249 (Homo sapiens)
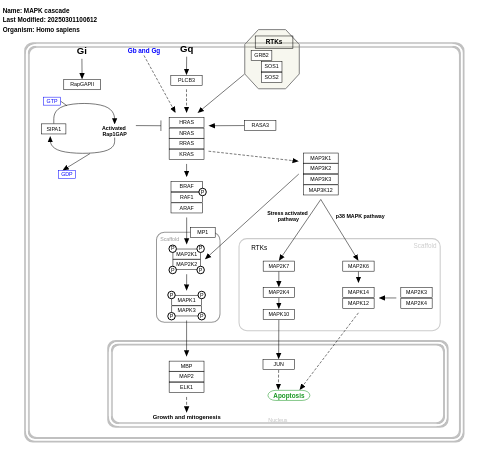
- MAPK cascade - WP422 (Homo sapiens)
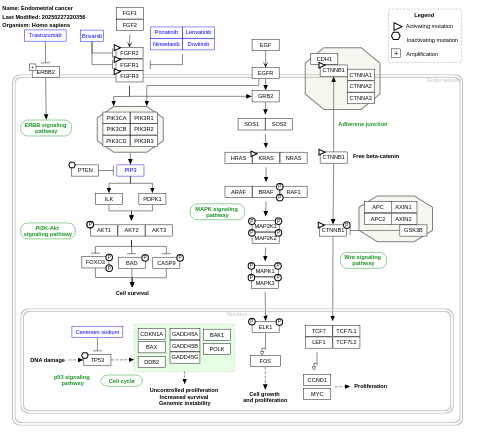
- Endometrial cancer - WP4155 (Homo sapiens)
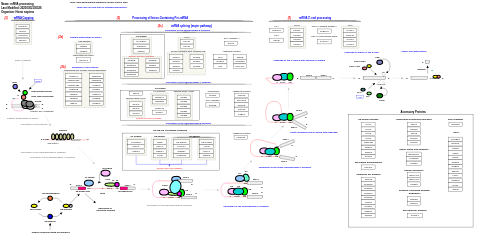
- mRNA processing - WP411 (Homo sapiens)
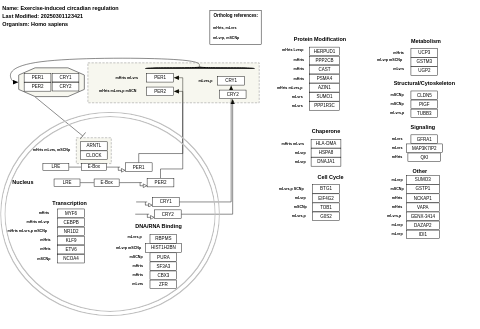
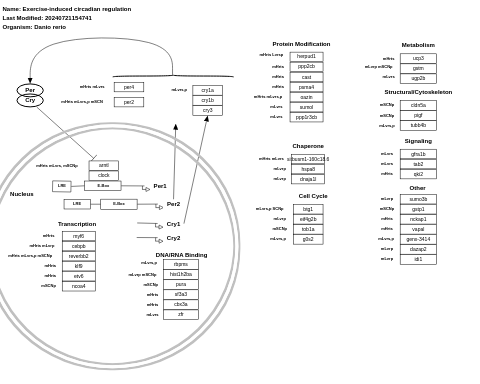
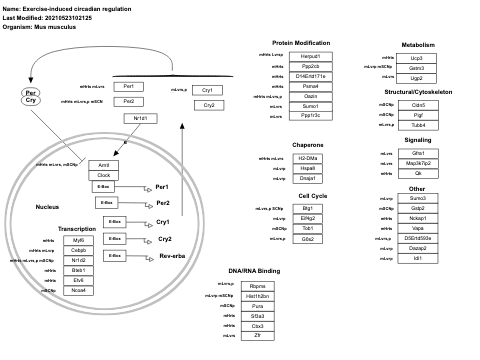
- Exercise-induced circadian regulation - WP410 (Homo sapiens)
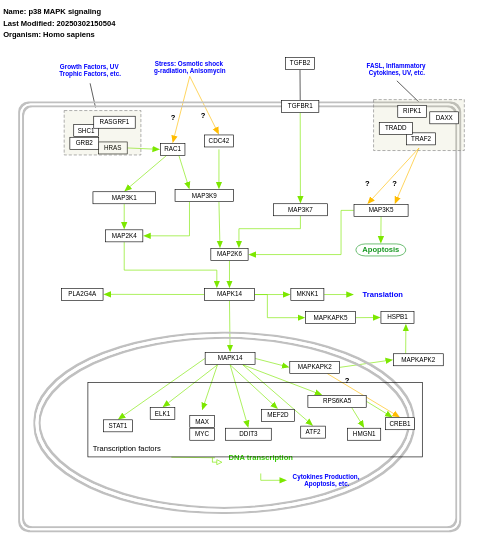
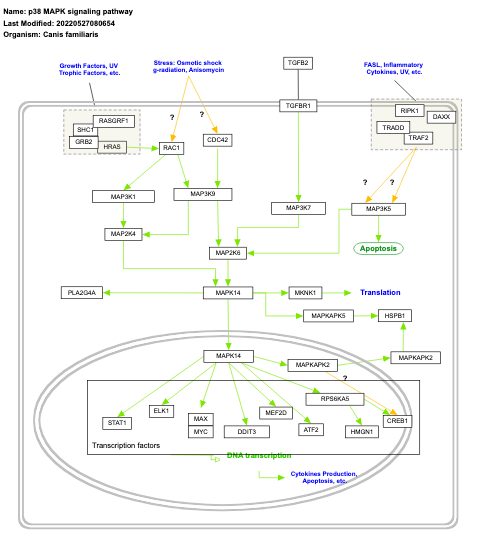
- p38 MAPK signaling - WP400 (Homo sapiens)
- H19, Rb-E2F1, and CDK-beta-catenin in colorectal cancer - WP3969 (Homo sapiens)
- TYROBP causal network in microglia - WP3945 (Homo sapiens)
- Microglia pathogen phagocytosis pathway - WP3937 (Homo sapiens)
- Apoptosis regulation by parathyroid hormone-related protein - WP3872 (Homo sapiens)
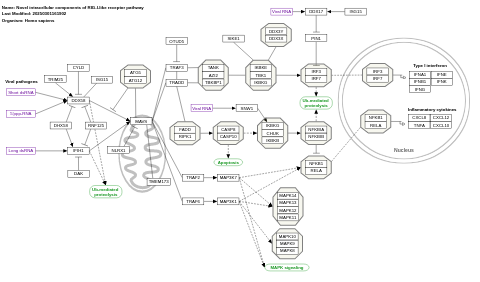
- Novel intracellular components of RIG-I-like receptor pathway - WP3865 (Homo sapiens)
- TLR4 signaling and tolerance - WP3851 (Homo sapiens)
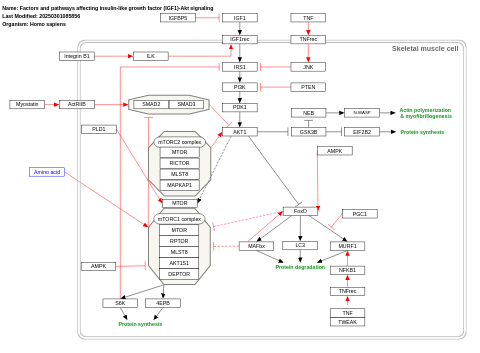
- Factors and pathways affecting insulin-like growth factor (IGF1)-Akt signaling - WP3850 (Homo sapiens)
- PI3K-AKT-mTOR signaling and therapeutic opportunities in prostate cancer - WP3844 (Homo sapiens)
- lncRNA-mediated mechanisms of therapeutic resistance - WP3672 (Homo sapiens)
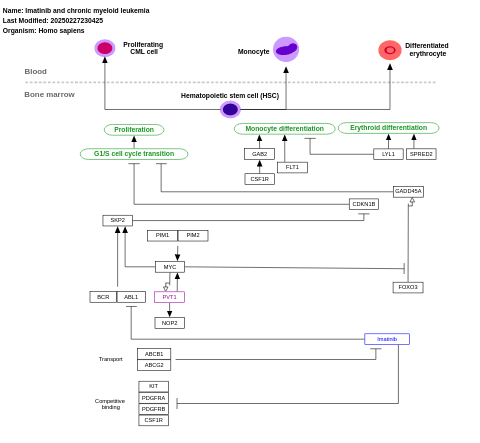
- Imatinib and chronic myeloid leukemia - WP3640 (Homo sapiens)
- Photodynamic therapy-induced NF-kB survival signaling - WP3617 (Homo sapiens)
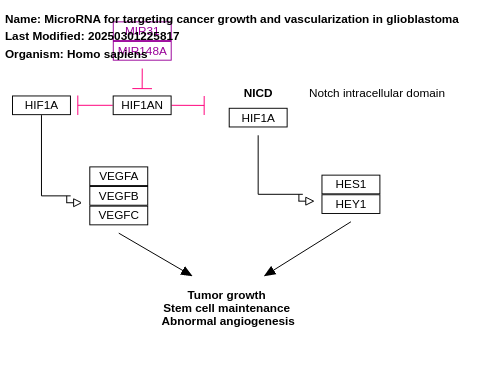
- MicroRNA for targeting cancer growth and vascularization in glioblastoma - WP3593 (Homo sapiens)
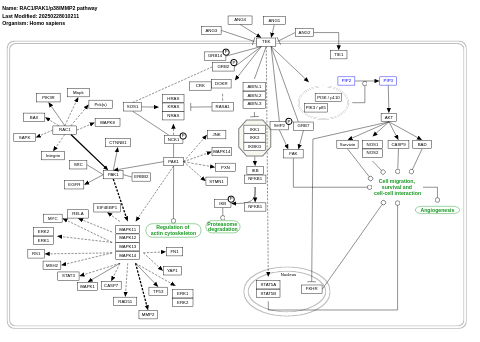
- RAC1/PAK1/p38/MMP2 pathway - WP3303 (Homo sapiens)
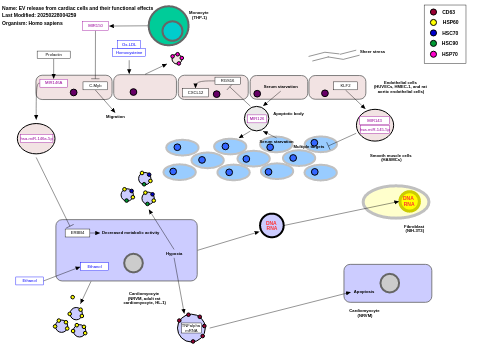
- EV release from cardiac cells and their functional effects - WP3297 (Homo sapiens)
- Glycogen metabolism - WP317 (Mus musculus)
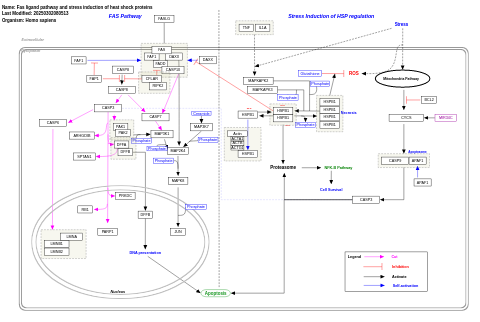
- Fas ligand pathway and stress induction of heat shock proteins - WP314 (Homo sapiens)
- mRNA processing - WP310 (Mus musculus)
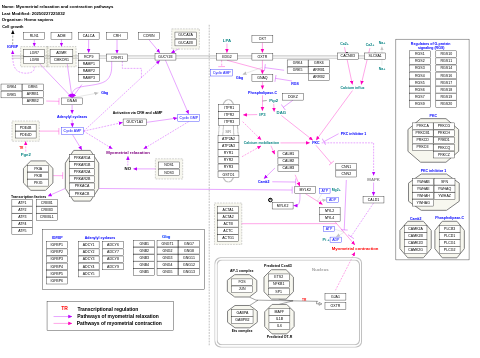
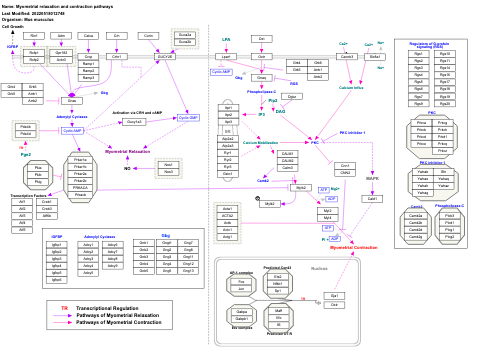
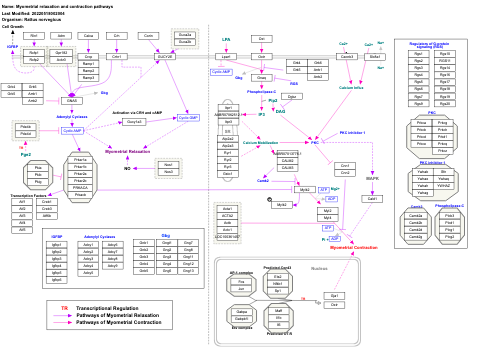
- Myometrial relaxation and contraction pathways - WP289 (Homo sapiens)
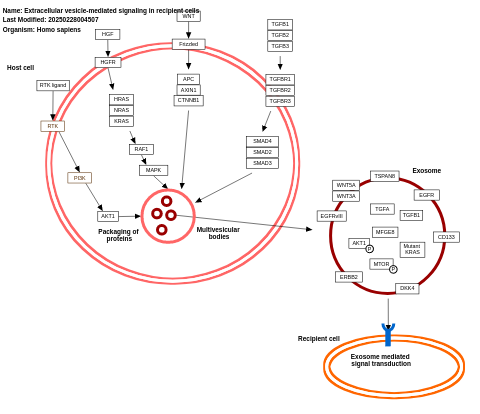
- Extracellular vesicle-mediated signaling in recipient cells - WP2870 (Homo sapiens)
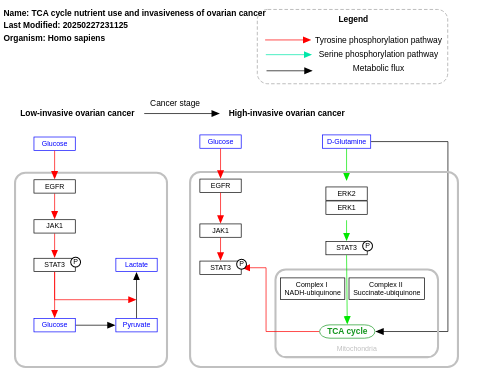
- TCA cycle nutrient use and invasiveness of ovarian cancer - WP2868 (Homo sapiens)
- Notch signaling - WP268 (Homo sapiens)
- Apoptosis - WP254 (Homo sapiens)
- Small ligand GPCRs - WP247 (Homo sapiens)
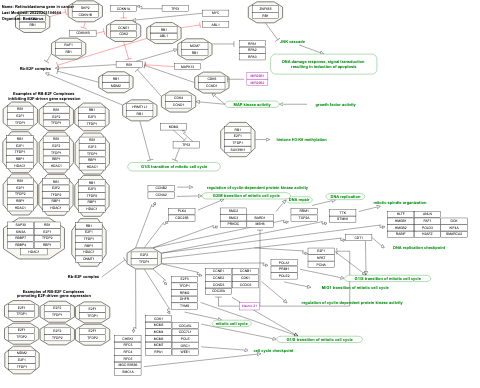
- Retinoblastoma gene in cancer - WP2446 (Homo sapiens)
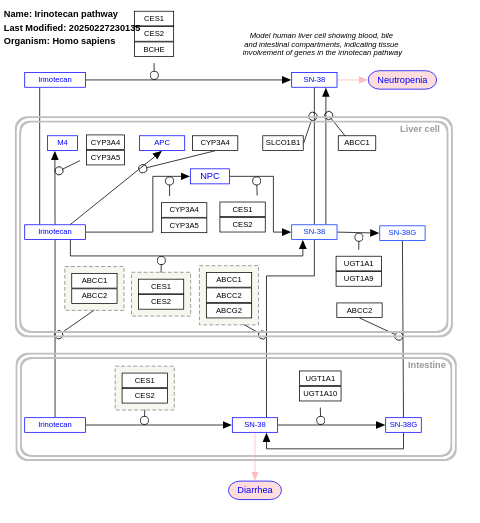
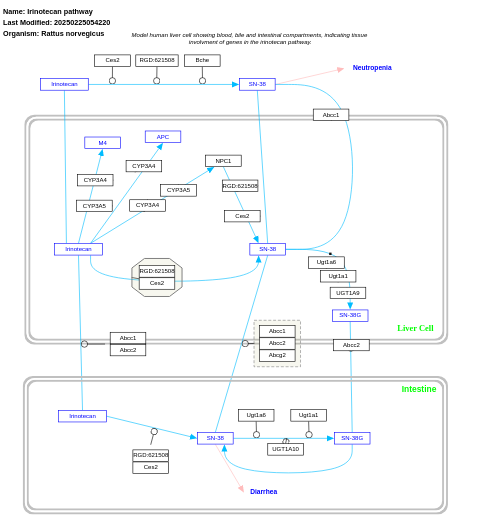
- Irinotecan pathway - WP229 (Homo sapiens)
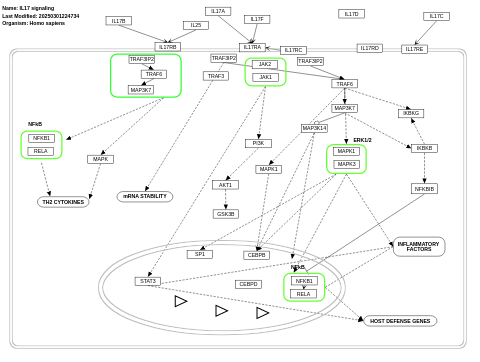
- IL17 signaling - WP2112 (Homo sapiens)
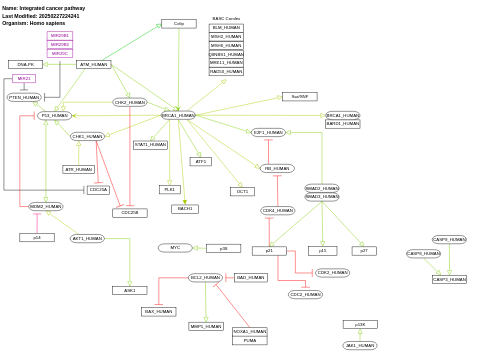
- Integrated cancer pathway - WP1971 (Homo sapiens)
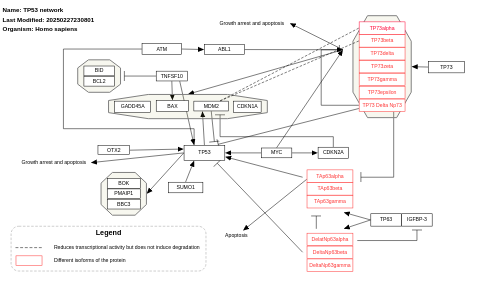
- TP53 network - WP1742 (Homo sapiens)
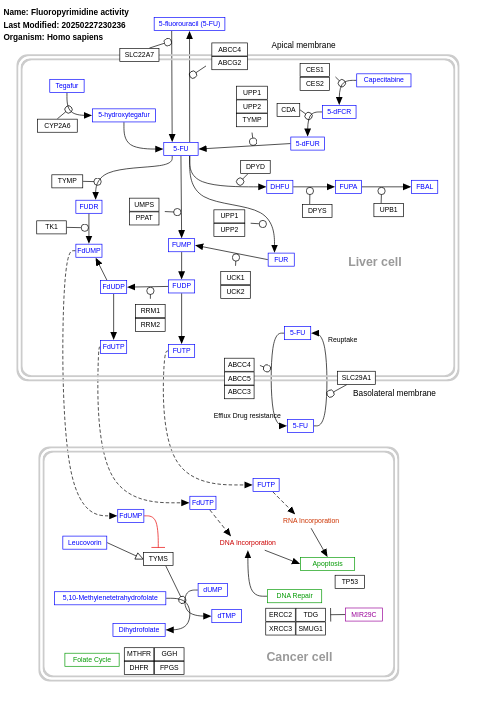
- Fluoropyrimidine activity - WP1601 (Homo sapiens)
- Glycolysis and gluconeogenesis - WP157 (Mus musculus)
- Biogenic amine synthesis - WP154 (Danio rerio)
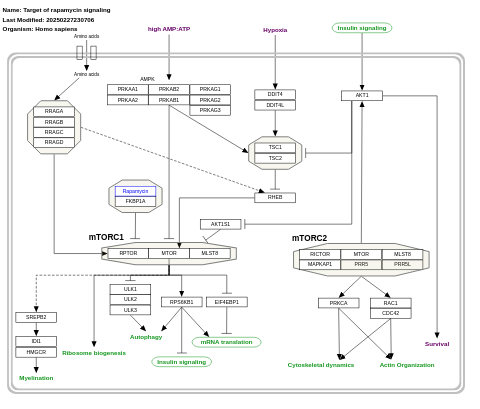
- Target of rapamycin signaling - WP1471 (Homo sapiens)
- Statin pathway - WP145 (Rattus norvegicus)
- Fatty acid beta-oxidation - WP143 (Homo sapiens)
- Androgen receptor signaling - WP138 (Homo sapiens)
- Pentose phosphate metabolism - WP134 (Homo sapiens)
- GPCRs, other - WP117 (Homo sapiens)
- B cell receptor signaling pathway - WP1025 (Bos taurus)
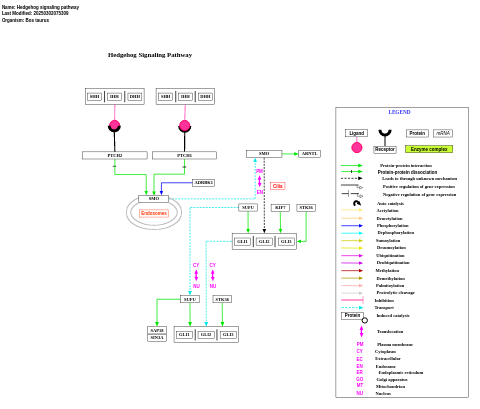
- Hedgehog signaling pathway - WP1022 (Bos taurus)
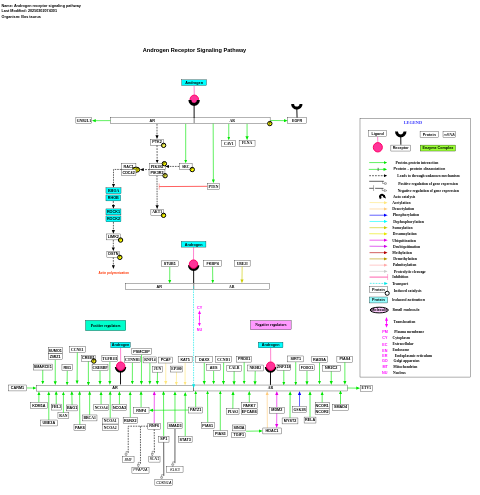
- Androgen receptor signaling pathway - WP1014 (Bos taurus)
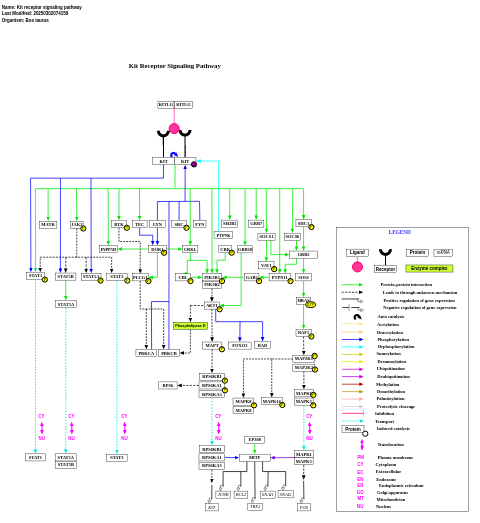
- Kit receptor signaling pathway - WP1004 (Bos taurus)
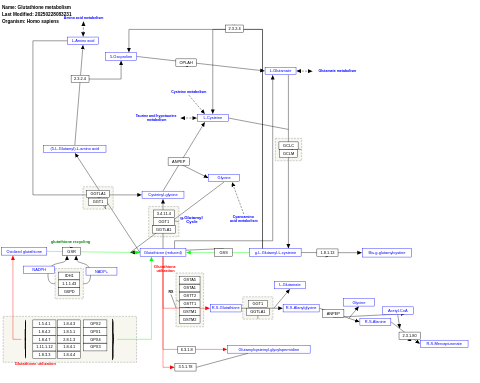
- Glutathione metabolism - WP100 (Homo sapiens)
- Glycogen metabolism - WP955 (Pan troglodytes)
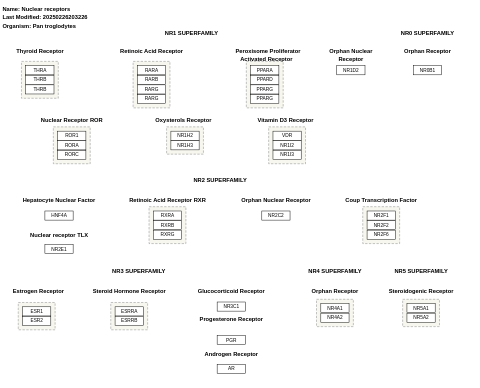
- Nuclear receptors - WP950 (Pan troglodytes)
- Nuclear receptors - WP831 (Gallus gallus)
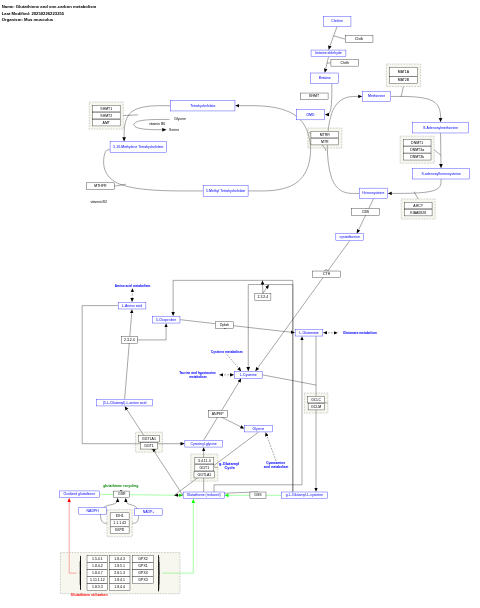
- Glutathione and one-carbon metabolism - WP730 (Mus musculus)
- DNA damage response (only ATM dependent) - WP710 (Homo sapiens)
- Aflatoxin B1 metabolism - WP699 (Homo sapiens)
- Integrin-mediated cell adhesion - WP6 (Mus musculus)
- Interferon type I signaling - WP585 (Homo sapiens)
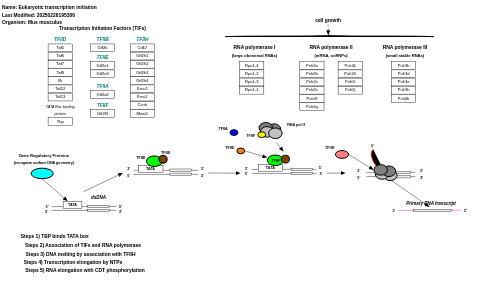
- Eukaryotic transcription initiation - WP567 (Mus musculus)
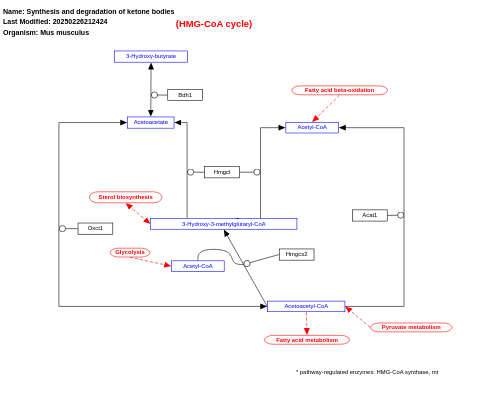
- Synthesis and degradation of ketone bodies - WP543 (Mus musculus)
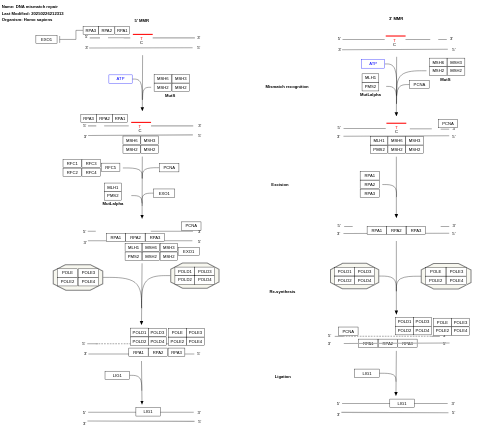
- DNA mismatch repair - WP531 (Homo sapiens)
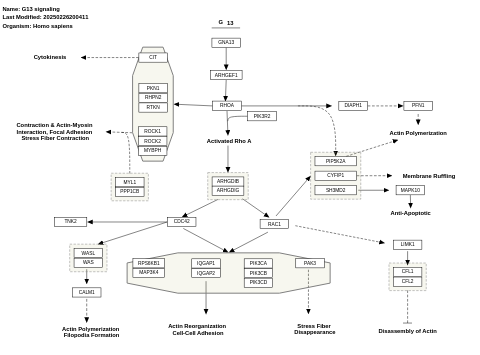
- G13 signaling - WP524 (Homo sapiens)
- Nuclear Receptors - WP509 (Mus musculus)
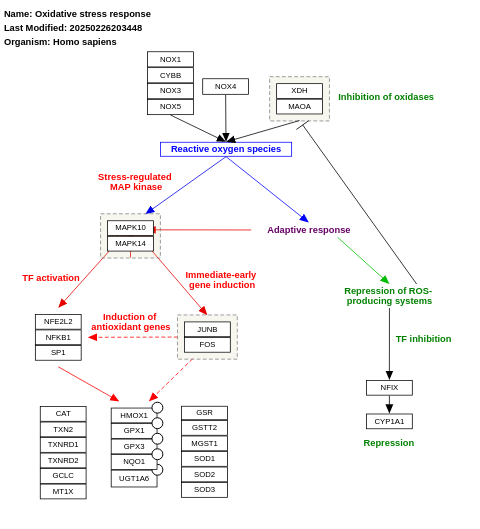
- Oxidative stress response - WP408 (Homo sapiens)
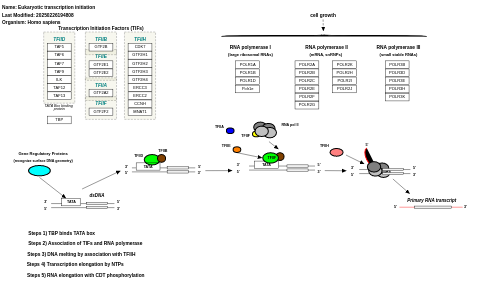
- Eukaryotic transcription initiation - WP405 (Homo sapiens)
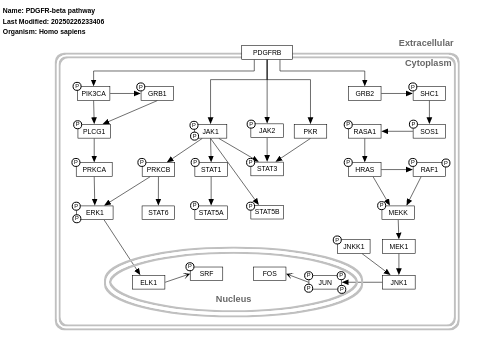
- PDGFR-beta pathway - WP3972 (Homo sapiens)
- Transcriptional activation by NRF2 in response to phytochemicals - WP3 (Homo sapiens)
- Nuclear receptors - WP217 (Rattus norvegicus)
- Homologous recombination - WP186 (Homo sapiens)
- Integrin-mediated cell adhesion - WP185 (Homo sapiens)
- Nuclear receptors - WP170 (Homo sapiens)
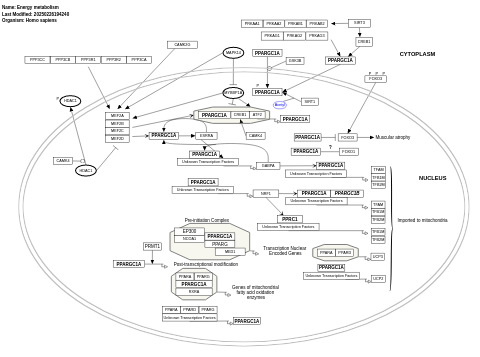
- Energy metabolism - WP1541 (Homo sapiens)
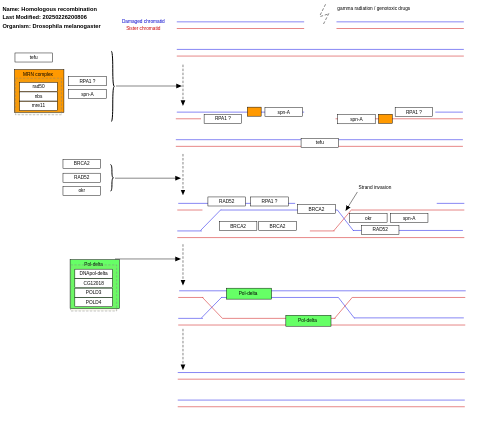
- Homologous recombination - WP1205 (Drosophila melanogaster)
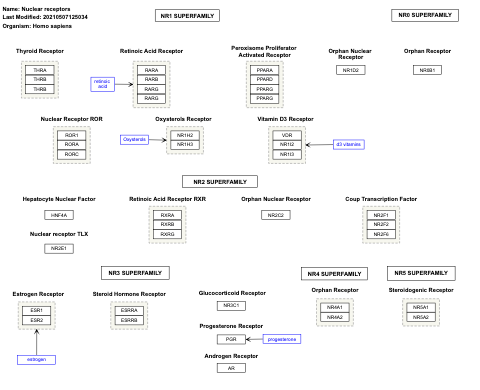
- Nuclear receptors - WP1184 (Canis familiaris)
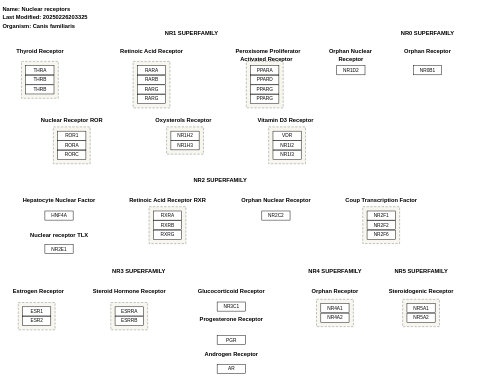
- Nuclear receptors - WP1068 (Bos taurus)
- HDAC6 interactions in the central nervous system - WP5426 (Homo sapiens)
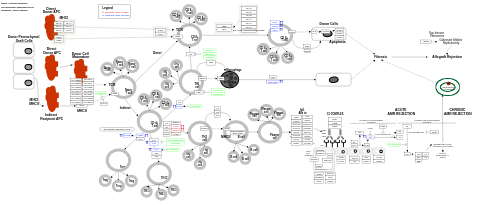
- Allograft rejection - WP2328 (Homo sapiens)
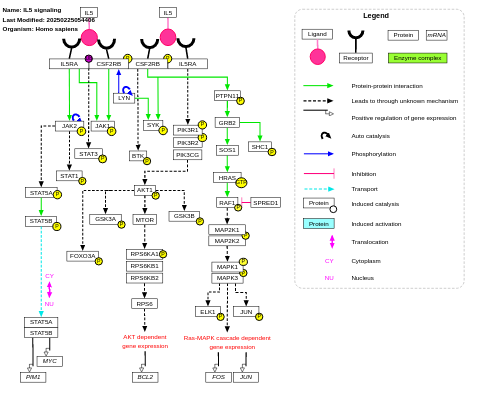
- IL5 signaling - WP127 (Homo sapiens)
- Irinotecan pathway - WP124 (Rattus norvegicus)
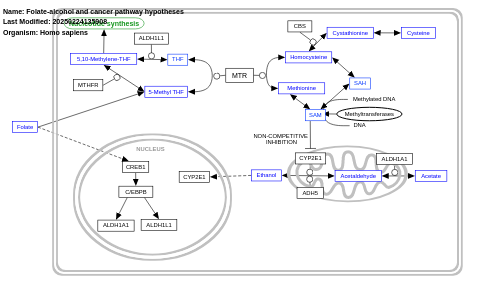
- Folate-alcohol and cancer pathway hypotheses - WP1589 (Homo sapiens)
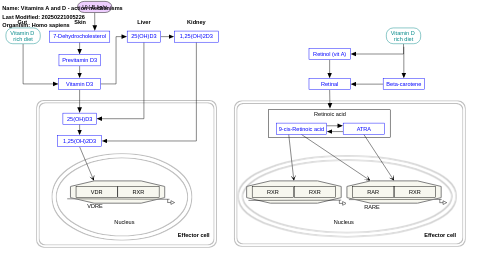
- Vitamins A and D - action mechanisms - WP4342 (Homo sapiens)
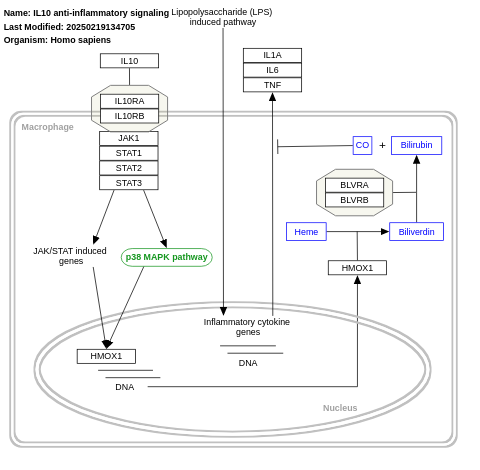
- IL10 anti-inflammatory signaling - WP4495 (Homo sapiens)
- IL26 signaling - WP5347 (Homo sapiens)
- Nicotine metabolism in liver cells - WP1600 (Homo sapiens)
- eIF5A regulation in response to inhibition of the nuclear export system - WP3302 (Homo sapiens)
- Octadecanoid formation from linoleic acid - WP5324 (Homo sapiens)
- Omega-3-fatty acids in senescence - WP5432 (Homo sapiens)
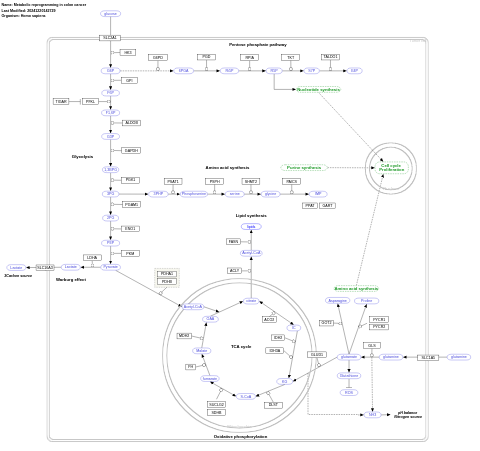
- Metabolic reprogramming in colon cancer - WP4290 (Homo sapiens)
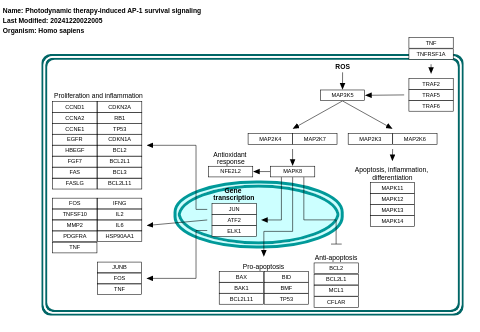
- Photodynamic therapy-induced AP-1 survival signaling - WP3611 (Homo sapiens)
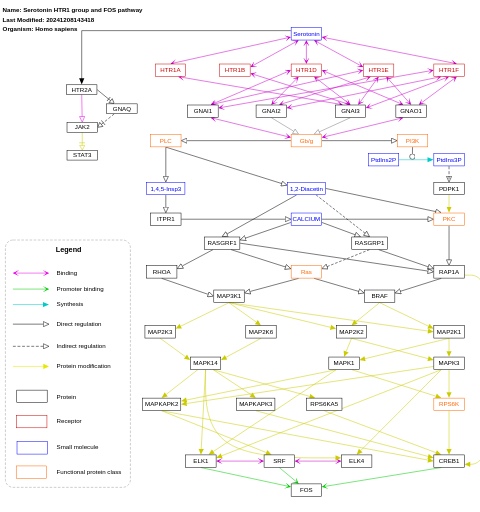
- Serotonin HTR1 group and FOS pathway - WP722 (Homo sapiens)
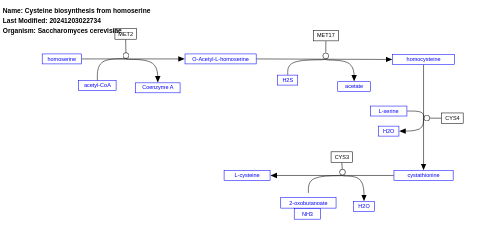
- Cysteine biosynthesis from homoserine - WP256 (Saccharomyces cerevisiae)
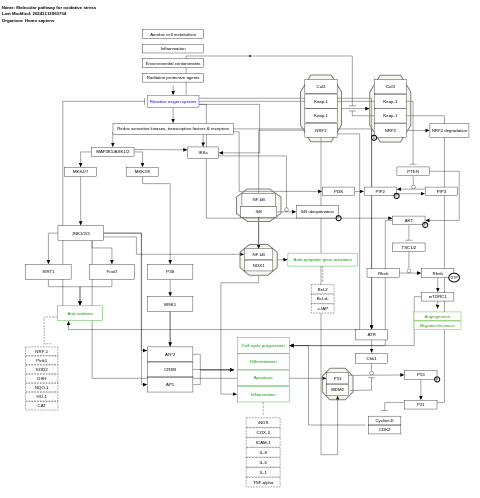
- Molecular pathway for oxidative stress - WP5477 (Homo sapiens)
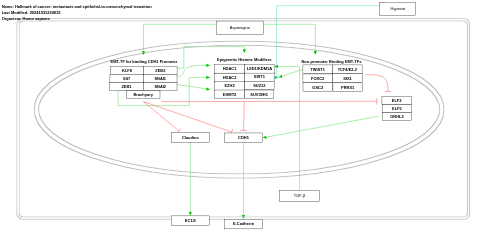
- Hallmark of cancer: metastasis and epithelial-to-mesenchymal transition - WP5469 (Homo sapiens)
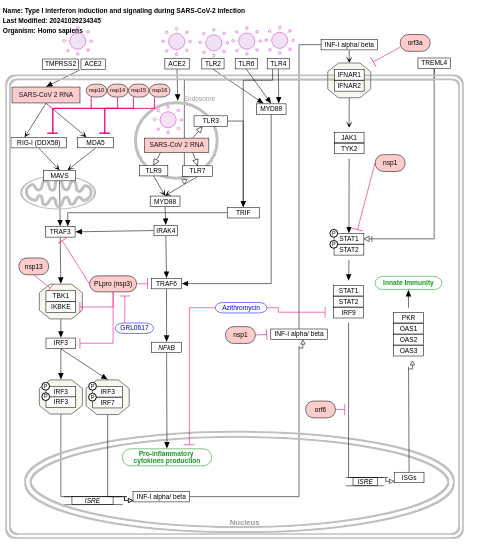
- Type I interferon induction and signaling during SARS-CoV-2 infection - WP4868 (Homo sapiens)
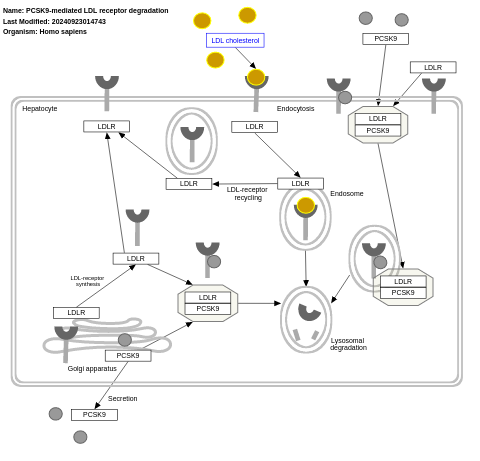
- PCSK9-mediated LDL receptor degradation - WP2846 (Homo sapiens)
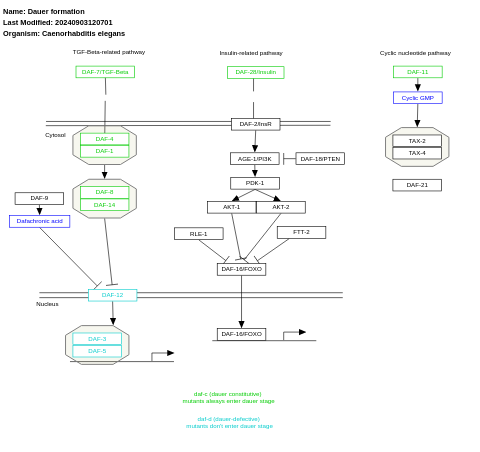
- Dauer formation - WP580 (Caenorhabditis elegans)
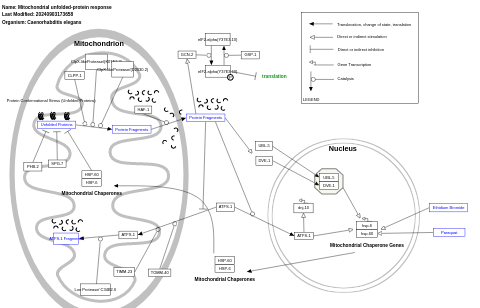
- Mitochondrial unfolded-protein response - WP525 (Caenorhabditis elegans)
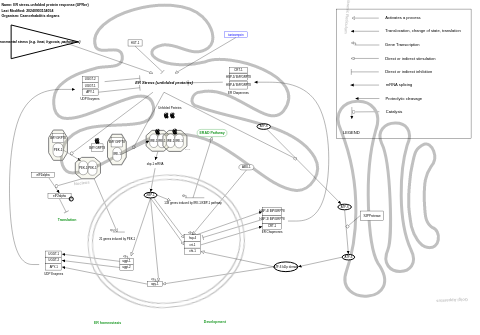
- ER stress-unfolded protein response (UPRer) - WP2578 (Caenorhabditis elegans)
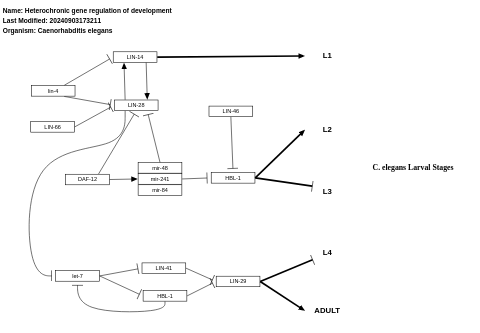
- Heterochronic gene regulation of development - WP2227 (Caenorhabditis elegans)
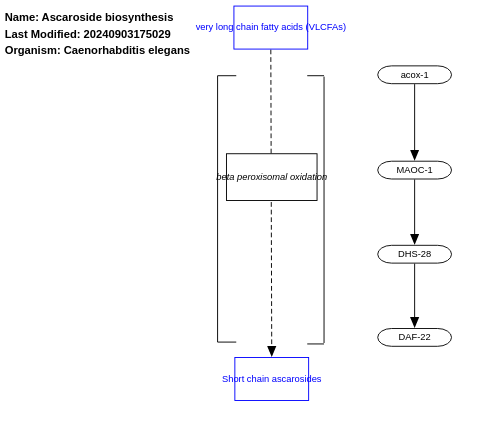
- Ascaroside biosynthesis - WP2224 (Caenorhabditis elegans)
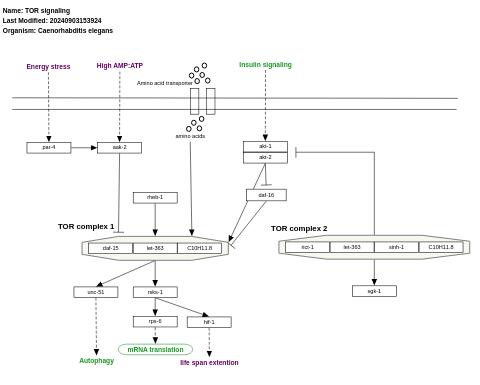
- TOR signaling - WP1489 (Caenorhabditis elegans)
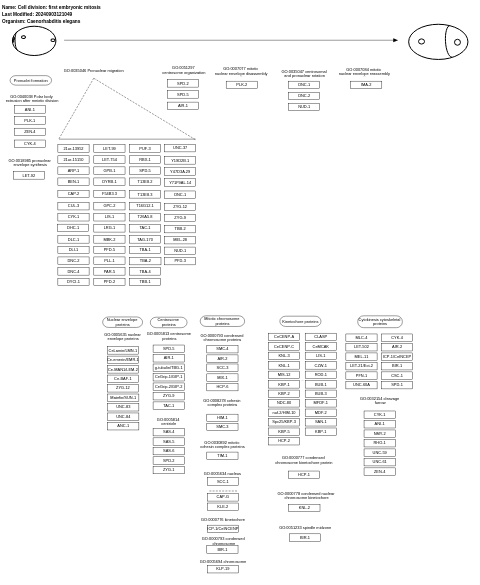
- Cell division: first embryonic mitosis - WP1411 (Caenorhabditis elegans)
- Biogenic amine synthesis - WP125 (Drosophila melanogaster)
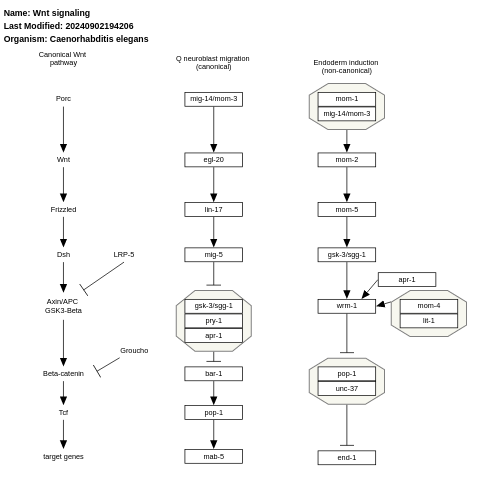
- Wnt signaling - WP235 (Caenorhabditis elegans)
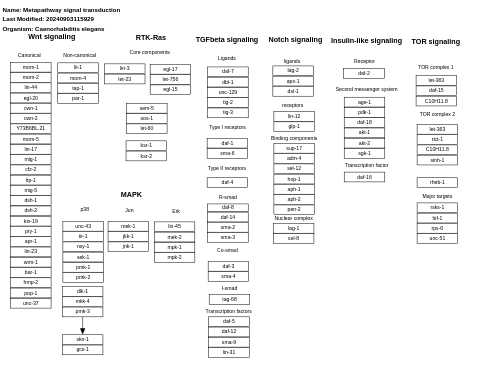
- Metapathway signal transduction - WP1546 (Caenorhabditis elegans)
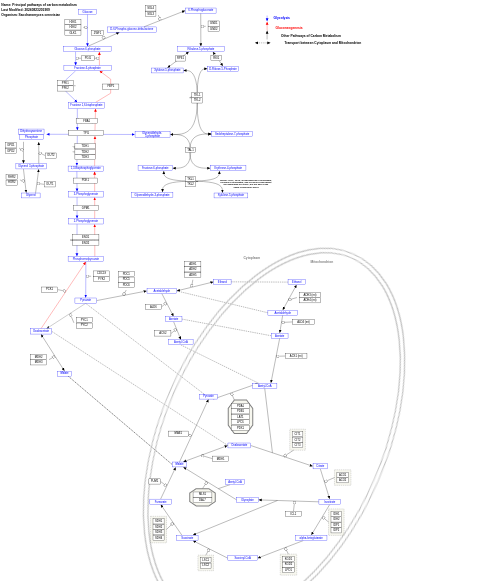
- Principal pathways of carbon metabolism - WP112 (Saccharomyces cerevisiae)
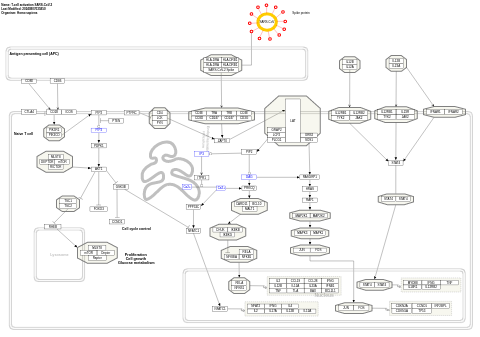
- T-cell activation SARS-CoV-2 - WP5098 (Homo sapiens)
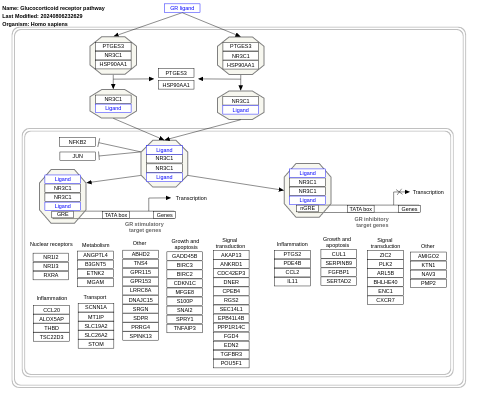
- Glucocorticoid receptor pathway - WP2880 (Homo sapiens)
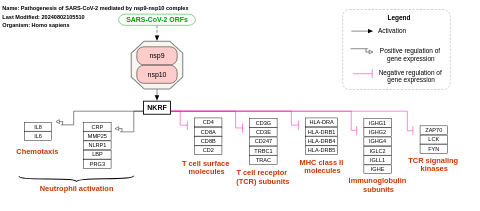
- Pathogenesis of SARS-CoV-2 mediated by nsp9-nsp10 complex - WP4884 (Homo sapiens)
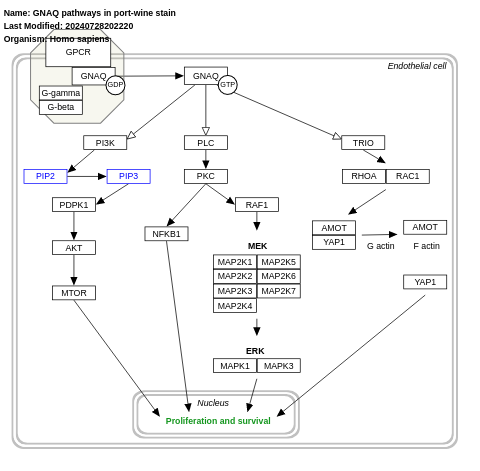
- GNAQ pathways in port-wine stain - WP5437 (Homo sapiens)
- Macrophage-stimulating protein (MSP) signaling - WP5353 (Homo sapiens)
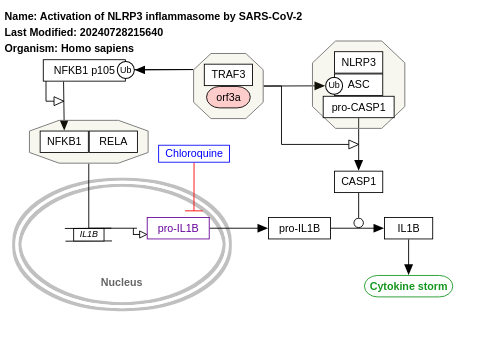
- Activation of NLRP3 inflammasome by SARS-CoV-2 - WP4876 (Homo sapiens)
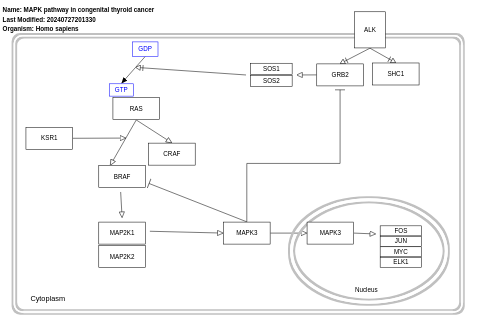
- MAPK pathway in congenital thyroid cancer - WP4928 (Homo sapiens)
- Docosahexaenoic acid oxylipin metabolism - WP5154 (Homo sapiens)
- Metabolism of alpha-linolenic acid - WP4586 (Homo sapiens)
- Blood clotting and drug effects - WP4580 (Homo sapiens)
- DYRK1A involvement regarding cell proliferation in brain development - WP5180 (Homo sapiens)
- 15q25 copy number variation - WP5408 (Homo sapiens)
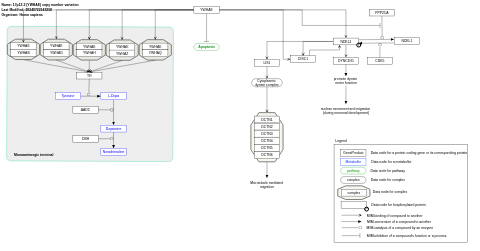
- 17p13.3 (YWHAE) copy number variation - WP5376 (Homo sapiens)
- Neurogenesis regulation in the olfactory epithelium - WP5265 (Homo sapiens)
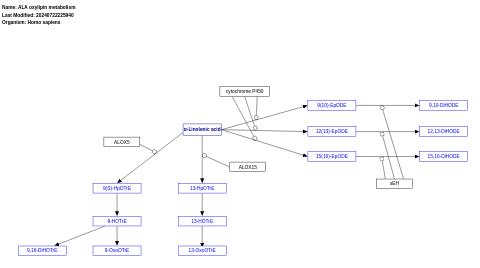
- ALA oxylipin metabolism - WP5136 (Homo sapiens)
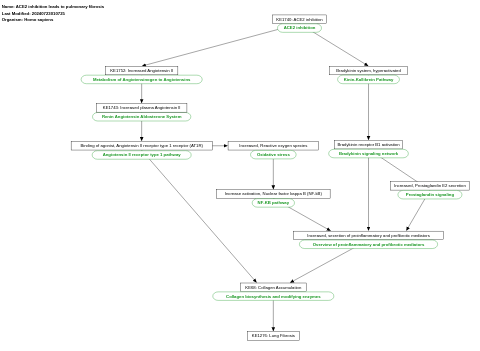
- ACE2 inhibition leads to pulmonary fibrosis - WP5035 (Homo sapiens)
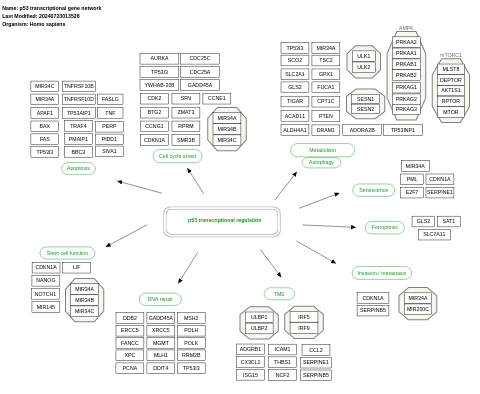
- p53 transcriptional gene network - WP4963 (Homo sapiens)
- EBV LMP1 signaling - WP984 (Bos taurus)
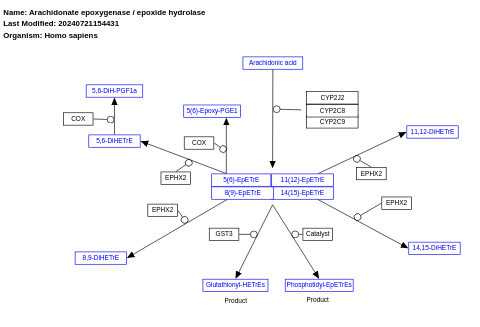
- Arachidonate epoxygenase / epoxide hydrolase - WP678 (Homo sapiens)
- Exercise-induced circadian regulation - WP562 (Danio rerio)
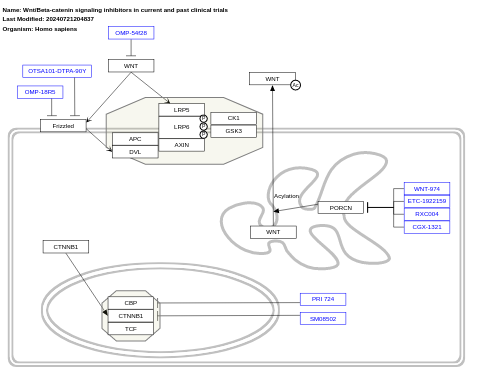
- Wnt/Beta-catenin signaling inhibitors in current and past clinical trials - WP5442 (Homo sapiens)
- 1p36 copy number variation syndrome - WP5345 (Homo sapiens)
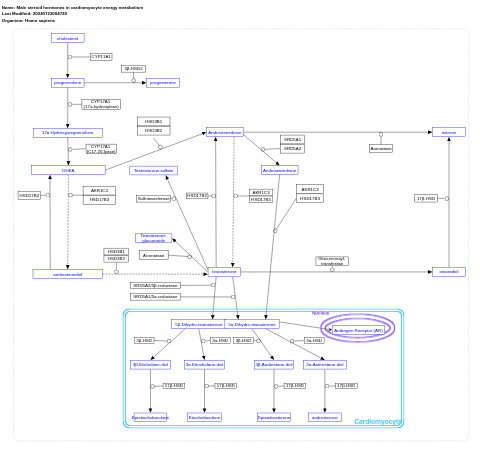
- Male steroid hormones in cardiomyocyte energy metabolism - WP5320 (Homo sapiens)
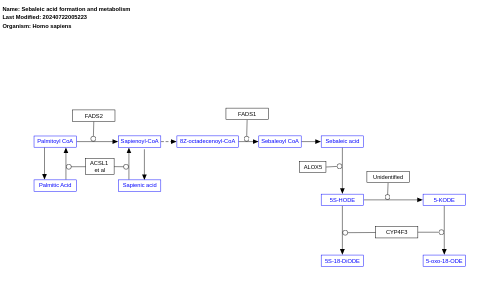
- Sebaleic acid formation and metabolism - WP5315 (Homo sapiens)
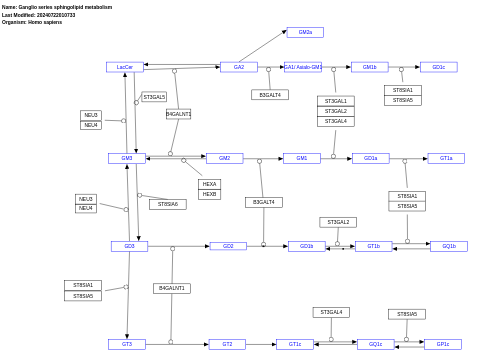
- Ganglio series sphingolipid metabolism - WP5299 (Homo sapiens)
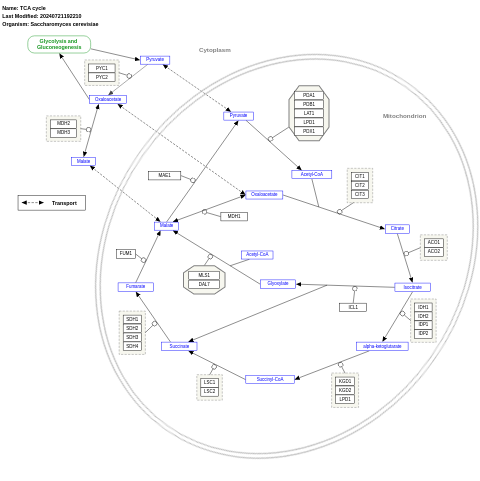
- TCA cycle - WP490 (Saccharomyces cerevisiae)
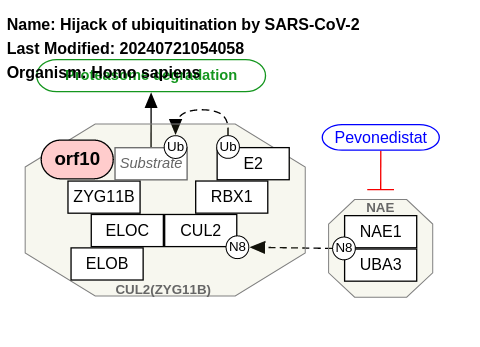
- Hijack of ubiquitination by SARS-CoV-2 - WP4860 (Homo sapiens)
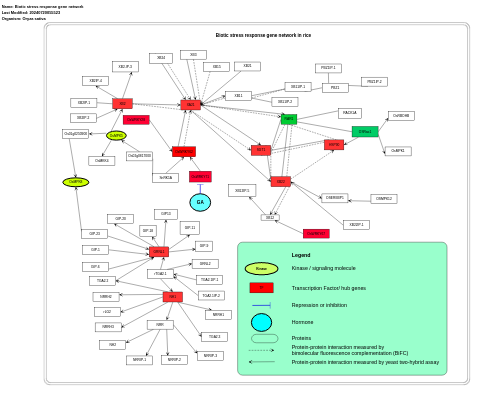
- Biotic stress response gene network - WP4364 (Oryza sativa)
- Dengue-2 interactions with complement and coagulation cascades - WP3896 (Homo sapiens)
- Nanomaterial-induced inflammasome activation - WP3890 (Homo sapiens)
- Quercetin and Nf-kB / AP-1 induced apoptosis - WP2435 (Homo sapiens)
- Nod-like receptor (NLR) signaling pathway - WP1294 (Rattus norvegicus)
- Nod-like receptor (NLR) signaling pathway - WP1256 (Mus musculus)
- Complement and coagulation cascades - WP1056 (Bos taurus)
- Nod-like receptor (NLR) signaling pathway - WP1032 (Bos taurus)
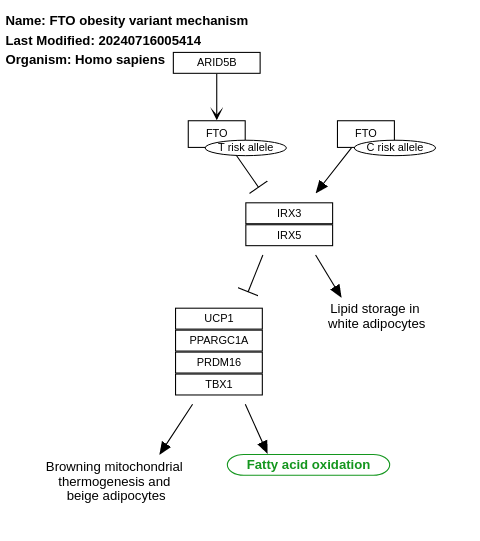
- FTO obesity variant mechanism - WP3407 (Homo sapiens)
- Sulfur amino acid biosynthesis - WP7 (Saccharomyces cerevisiae)
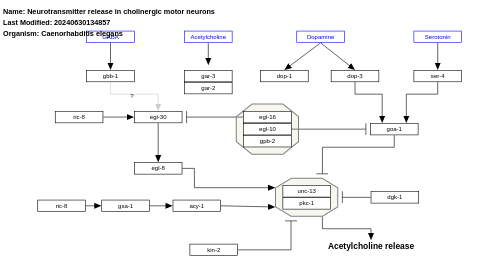
- Neurotransmitter release in cholinergic motor neurons - WP1401 (Caenorhabditis elegans)
- Hippo signaling regulation - WP4540 (Homo sapiens)
- Nod-like receptor (NLR) signaling - WP288 (Homo sapiens)
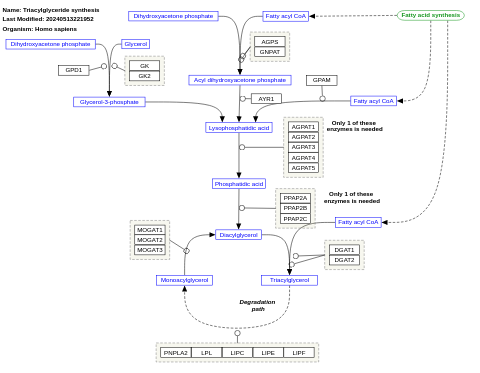
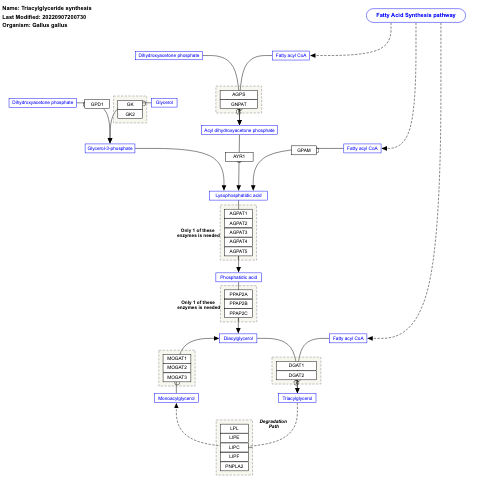
- Triacylglyceride synthesis - WP325 (Homo sapiens)
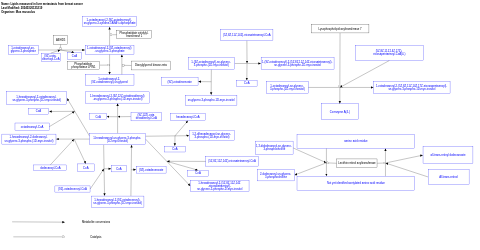
- Lipids measured in liver metastasis from breast cancer - WP4627 (Mus musculus)
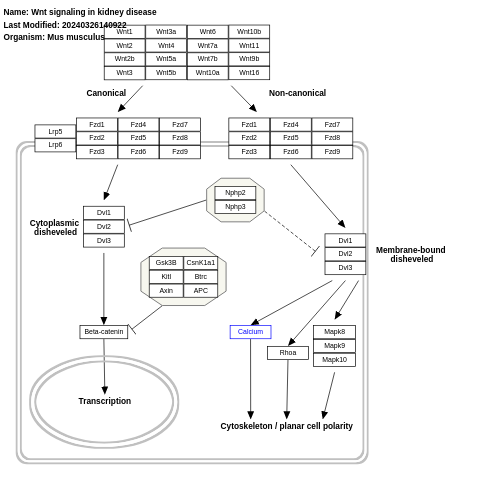
- Wnt signaling in kidney disease - WP3857 (Mus musculus)
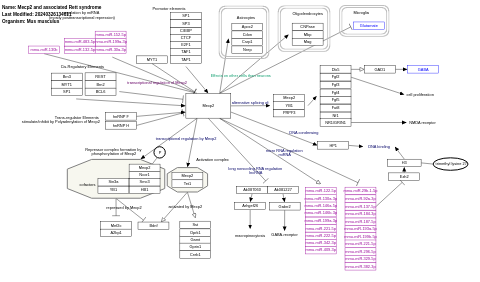
- Mecp2 and associated Rett syndrome - WP2910 (Mus musculus)
- Extrafollicular and follicular B cell activation by SARS-CoV-2 - WP5218 (Homo sapiens)
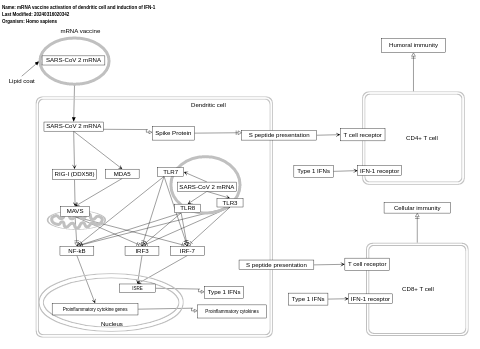
- mRNA vaccine activation of dendritic cell and induction of IFN-1 - WP5187 (Homo sapiens)
- Hematopoietic stem cell differentiation - WP2849 (Homo sapiens)
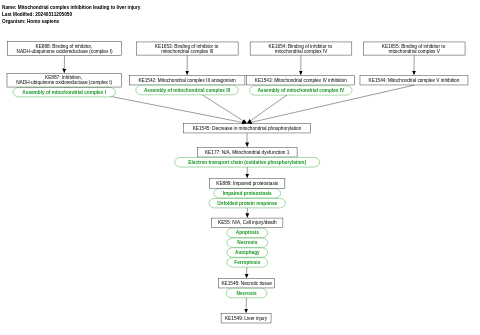
- Mitochondrial complex inhibition leading to liver injury - WP5034 (Homo sapiens)
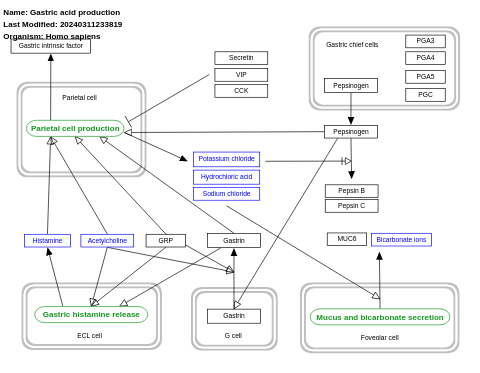
- Gastric acid production - WP2596 (Homo sapiens)
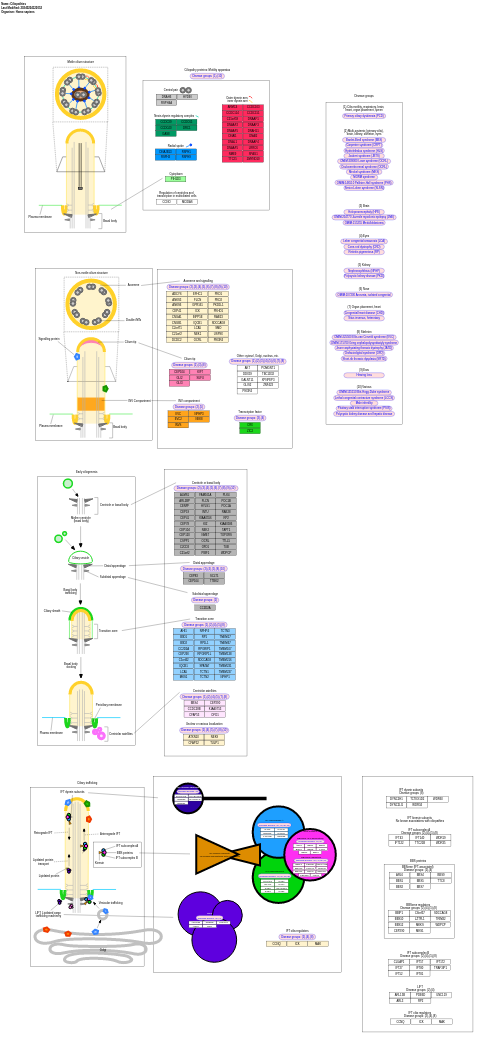
- Ciliopathies - WP4803 (Homo sapiens)
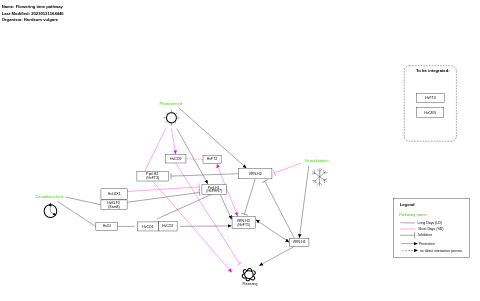
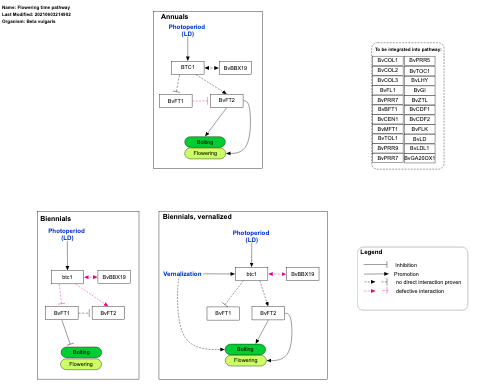
- Flowering time pathway - WP4339 (Brassica napus)
- Flowering time pathway - WP2625 (Hordeum vulgare)
- Evolocumab mechanism to reduce LDL cholesterol - WP3408 (Homo sapiens)
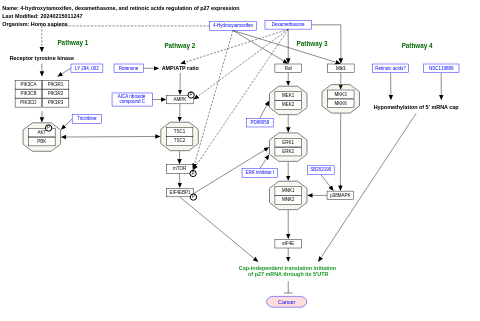
- 4-hydroxytamoxifen, dexamethasone, and retinoic acids regulation of p27 expression - WP3879 (Homo sapiens)
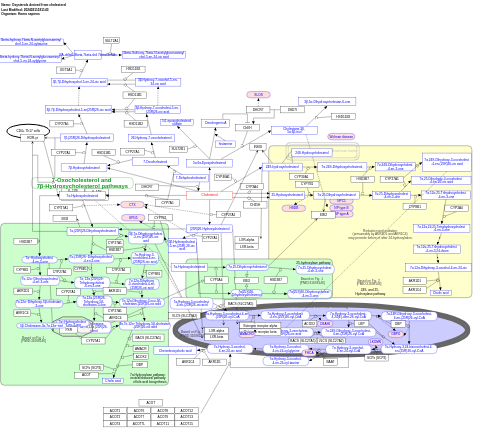
- Oxysterols derived from cholesterol - WP4545 (Homo sapiens)
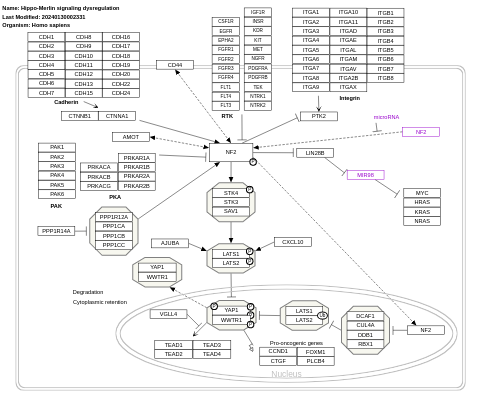
- Hippo-Merlin signaling dysregulation - WP4541 (Homo sapiens)
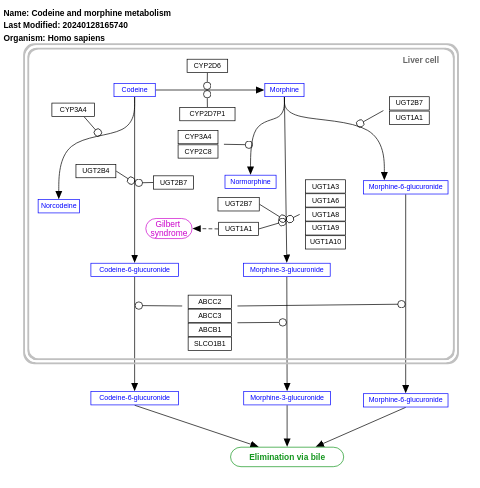
- Codeine and morphine metabolism - WP1604 (Homo sapiens)
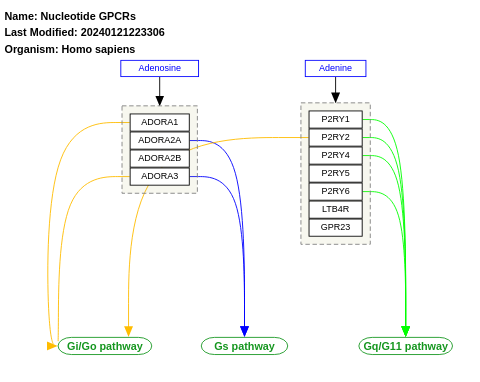
- Nucleotide GPCRs - WP80 (Homo sapiens)
- Tryptophan metabolism - WP79 (Mus musculus)
- Notch signaling pathway - WP29 (Mus musculus)
- T-cell antigen receptor (TCR) pathway during Staphylococcus aureus infection - WP3863 (Homo sapiens)
- Canonical and non-canonical Notch signaling - WP3845 (Homo sapiens)
- Purine metabolism - WP2185 (Mus musculus)
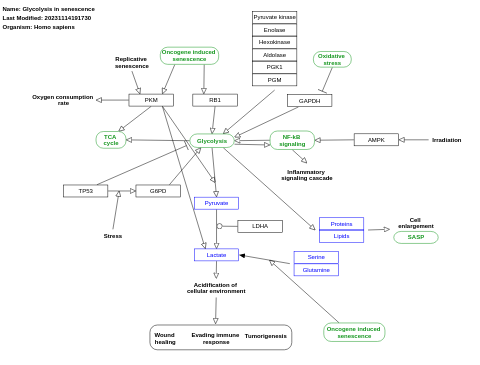
- Glycolysis in senescence - WP5049 (Homo sapiens)
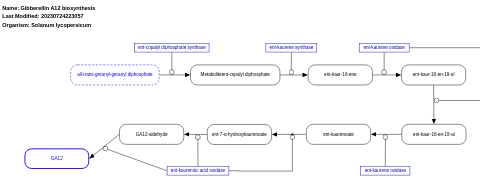
- Gibberellin A12 biosynthesis - WP2628 (Solanum lycopersicum)
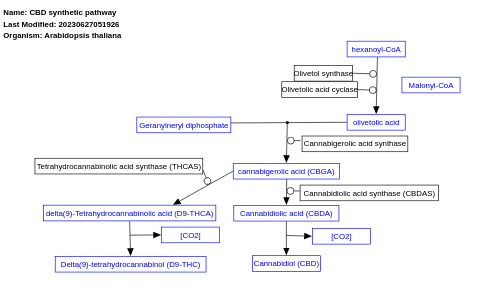
- CBD synthetic pathway - WP5392 (Arabidopsis thaliana)
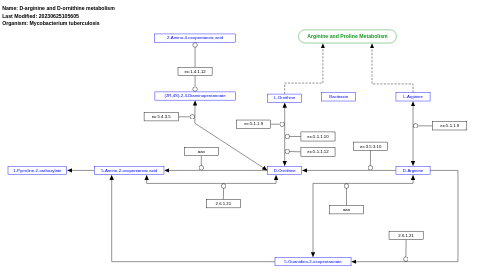
- D-arginine and D-ornithine metabolism - WP1642 (Mycobacterium tuberculosis)
- Osteoblast signaling - WP227 (Rattus norvegicus)
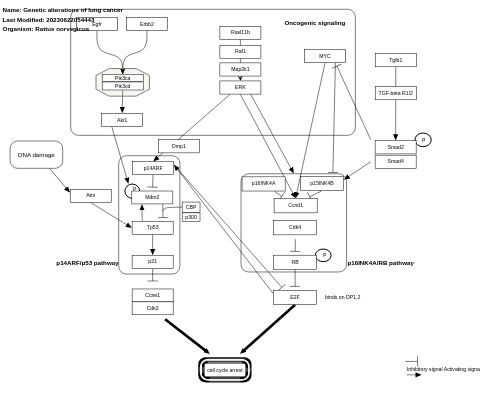
- Genetic alterations of lung cancer - WP1968 (Rattus norvegicus)
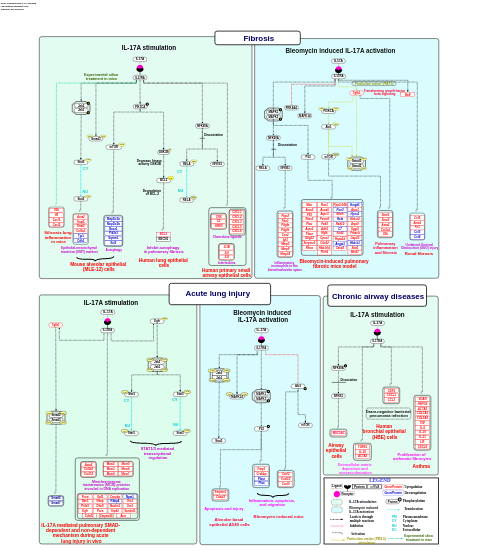
- Comprehensive IL-17A signaling - WP5242 (Mus musculus)
- Mechanisms associated with pluripotency - WP1763 (Mus musculus)
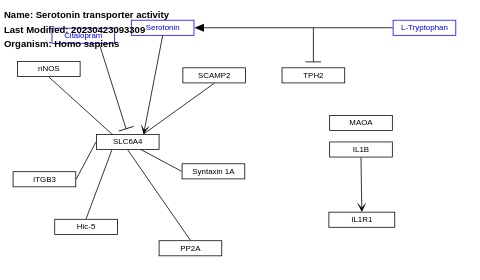
- Serotonin transporter activity - WP1455 (Homo sapiens)
- VEGFA-VEGFR2 signaling - WP3888 (Homo sapiens)
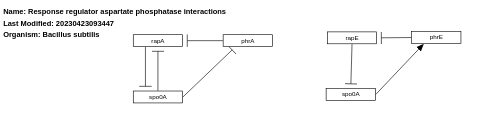
- Response regulator aspartate phosphatase interactions - WP1466 (Bacillus subtilis)
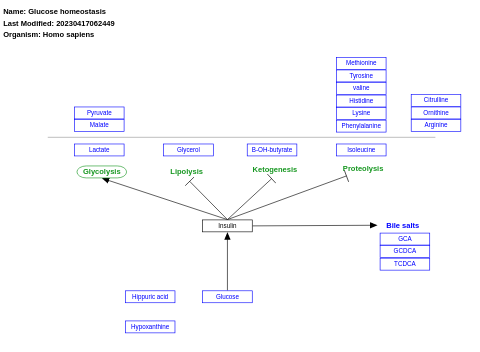
- Glucose homeostasis - WP661 (Homo sapiens)
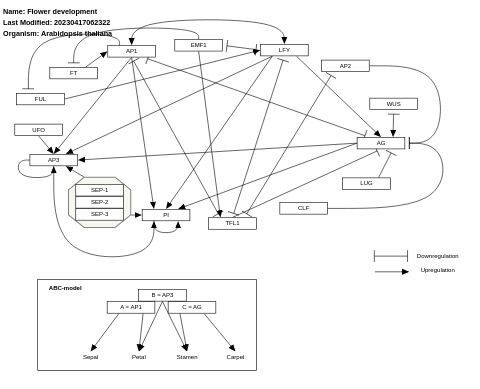
- Flower development - WP618 (Arabidopsis thaliana)
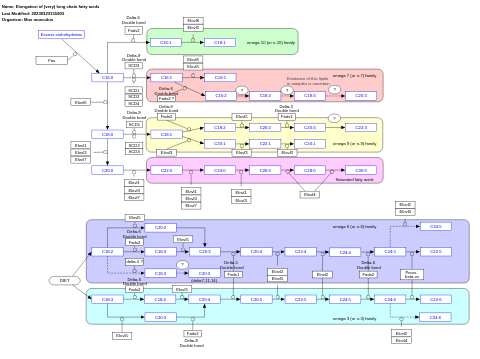
- Elongation of (very) long chain fatty acids - WP4491 (Mus musculus)
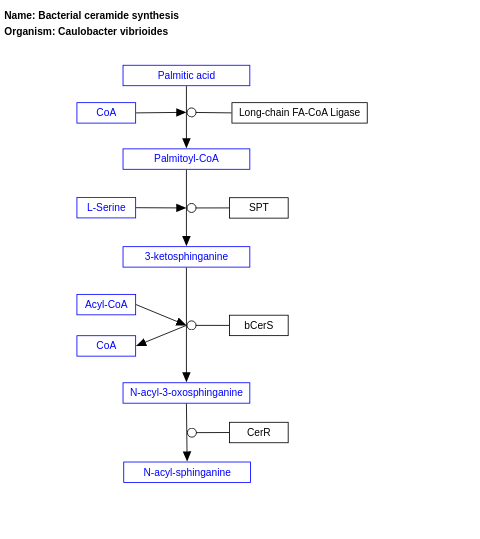
- Bacterial ceramide synthesis - WP5271 (Caulobacter vibrioides)
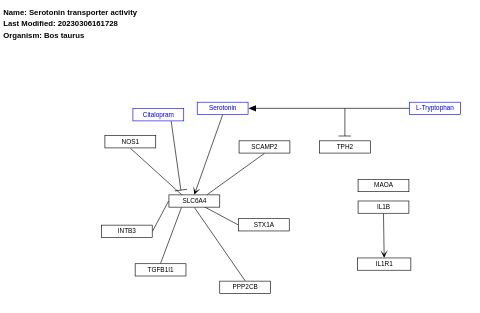
- Serotonin transporter activity - WP3138 (Bos taurus)
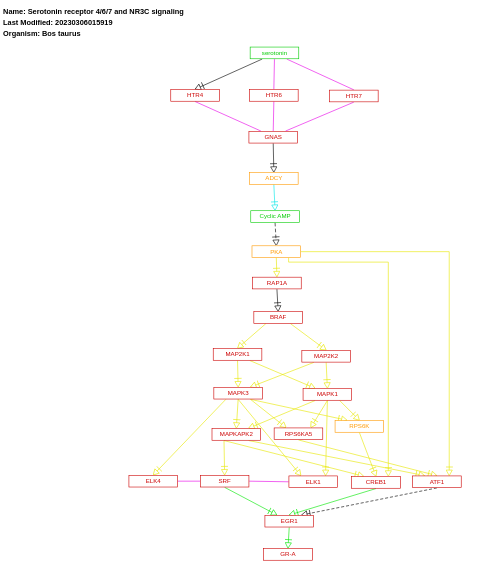
- Serotonin receptor 4/6/7 and NR3C signaling - WP3272 (Bos taurus)
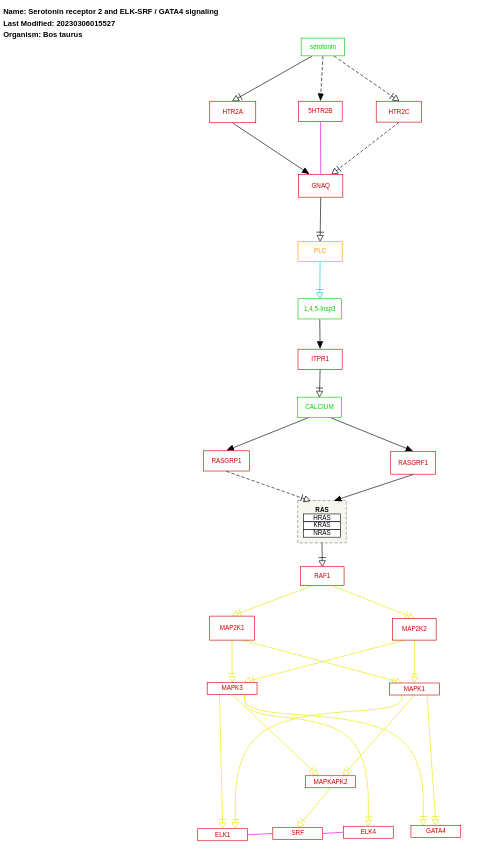
- Serotonin receptor 2 and ELK-SRF / GATA4 signaling - WP3242 (Bos taurus)
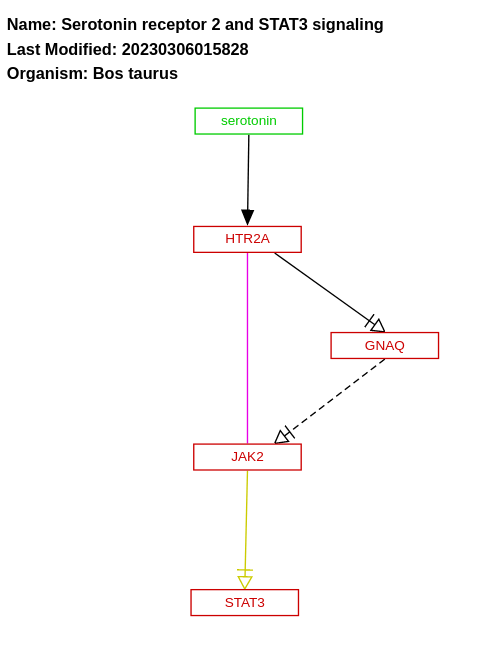
- Serotonin receptor 2 and STAT3 signaling - WP3115 (Bos taurus)
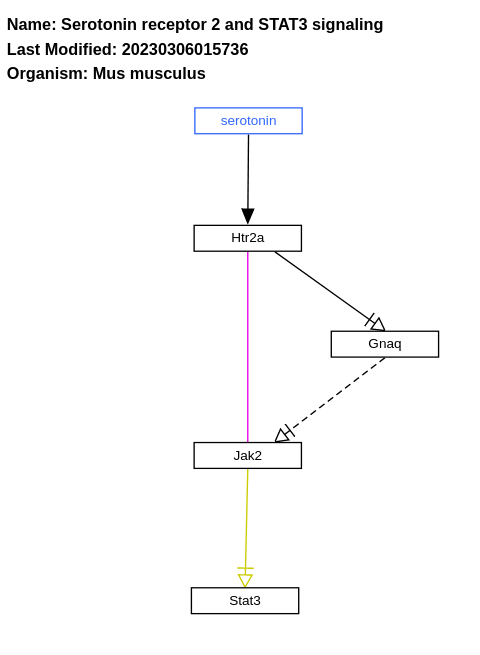
- Serotonin receptor 2 and STAT3 signaling - WP2079 (Mus musculus)
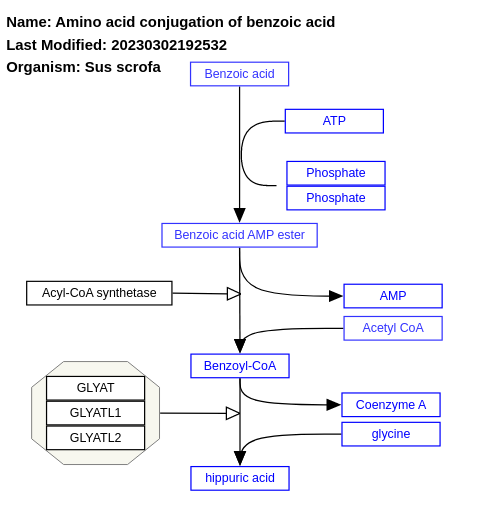
- Amino acid conjugation of benzoic acid - WP1577 (Sus scrofa)
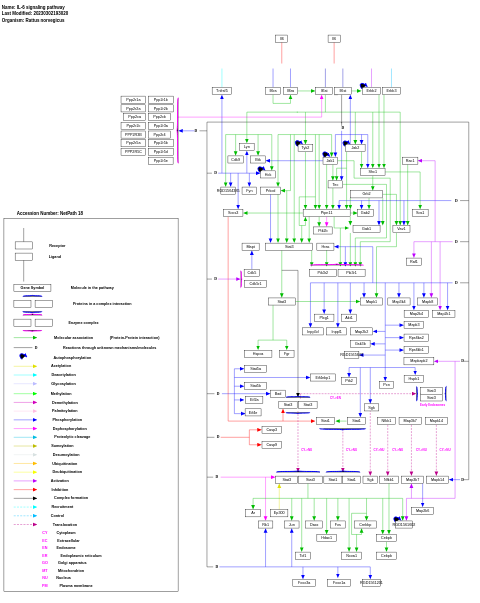
- IL-6 signaling pathway - WP135 (Rattus norvegicus)
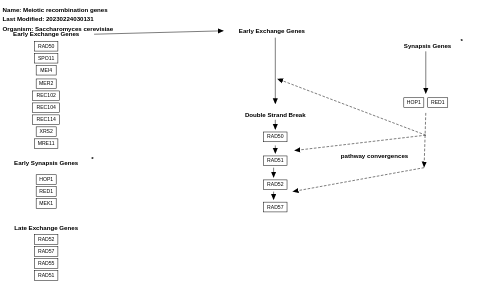
- Meiotic recombination genes - WP377 (Saccharomyces cerevisiae)
- GPCRs, class A rhodopsin-like - WP473 (Rattus norvegicus)
- Nuclear factor, erythroid-derived 2, like 2 signaling pathway - WP2376 (Rattus norvegicus)
- Metapathway biotransformation - WP1286 (Rattus norvegicus)
- Fatty acid biosynthesis - WP568 (Drosophila melanogaster)
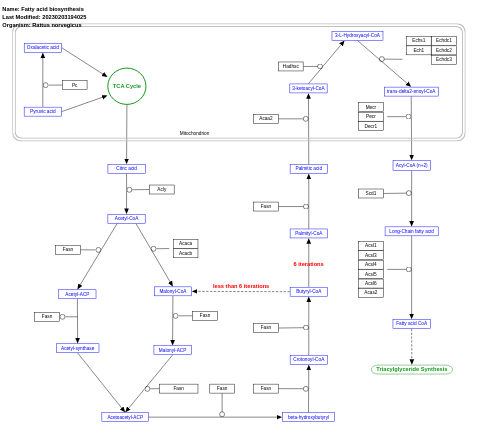
- Fatty acid biosynthesis - WP504 (Rattus norvegicus)
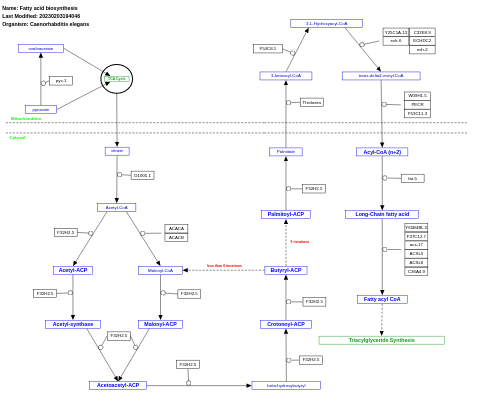
- Fatty acid biosynthesis - WP38 (Caenorhabditis elegans)
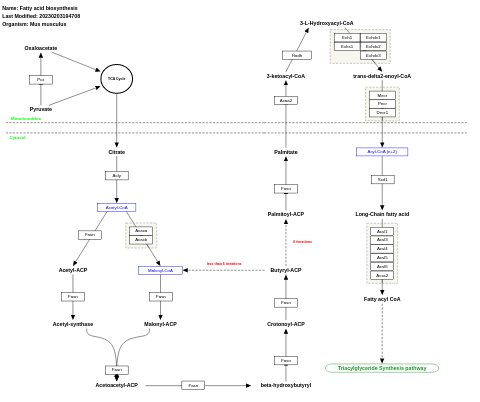
- Fatty acid biosynthesis - WP336 (Mus musculus)
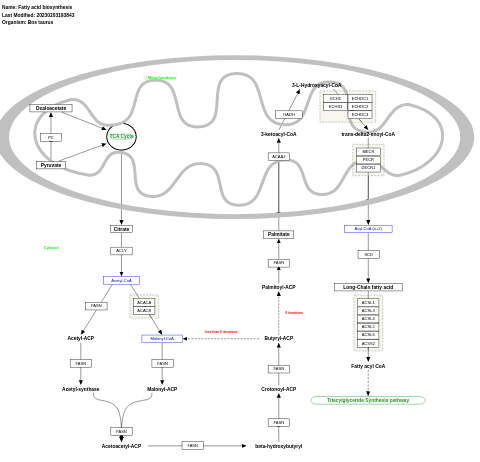
- Fatty acid biosynthesis - WP1020 (Bos taurus)
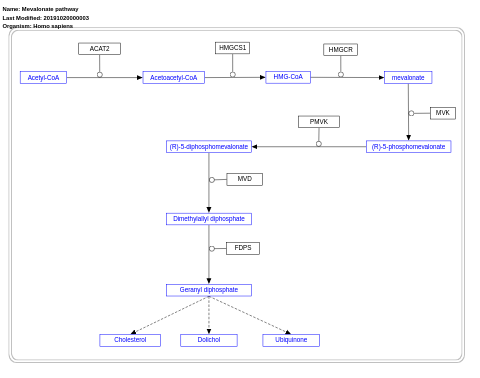
- Mevalonate pathway - WP3963 (Homo sapiens)
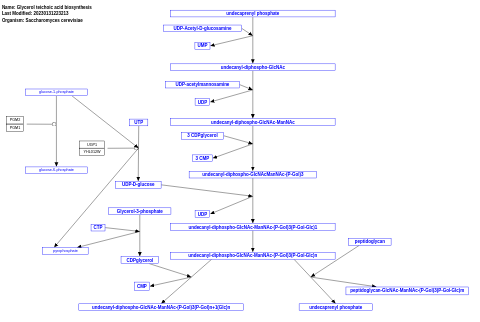
- Glycerol teichoic acid biosynthesis - WP563 (Saccharomyces cerevisiae)
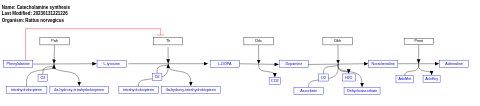
- Catecholamine synthesis - WP513 (Rattus norvegicus)
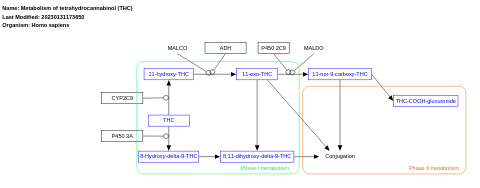
- Metabolism of tetrahydrocannabinol (THC) - WP4174 (Homo sapiens)
- Primary focal segmental glomerulosclerosis (FSGS) - WP3233 (Bos taurus)
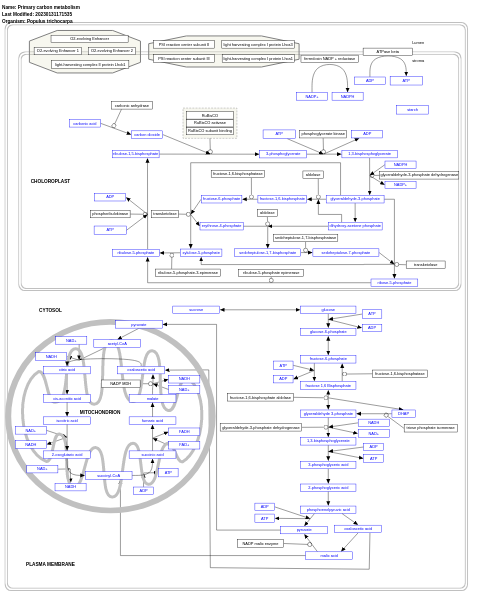
- Primary carbon metabolism - WP2859 (Populus trichocarpa)
- mRNA processing - WP123 (Caenorhabditis elegans)
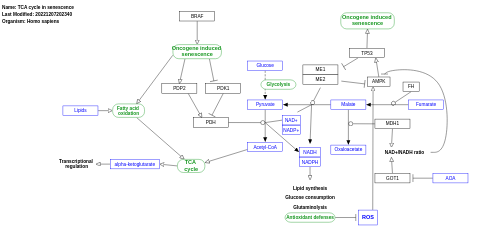
- TCA cycle in senescence - WP5050 (Homo sapiens)
- Retinol metabolism - WP1297 (Rattus norvegicus)
- Retinol metabolism - WP1259 (Mus musculus)
- Triacylglyceride synthesis - WP782 (Gallus gallus)
- mRNA processing - WP467 (Danio rerio)
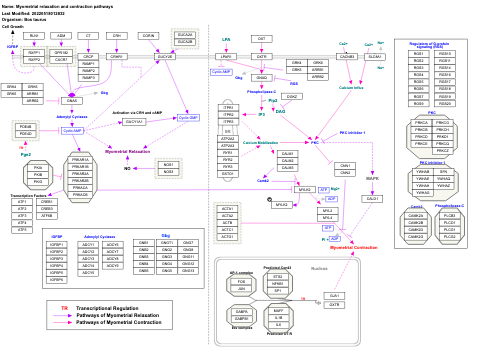
- Myometrial relaxation and contraction pathways - WP385 (Mus musculus)
- Myometrial relaxation and contraction pathways - WP3276 (Bos taurus)
- Semaphorin interactions - WP3151 (Bos taurus)
- mRNA processing - WP142 (Drosophila melanogaster)
- Myometrial relaxation and contraction pathways - WP140 (Rattus norvegicus)
- mRNA processing - WP1229 (Anopheles gambiae)
- mRNA Processing - WP1142 (Canis familiaris)
- Retinoblastoma gene in cancer - WP3206 (Bos taurus)
- Glycolysis and gluconeogenesis - WP1567 (Mycobacterium tuberculosis)
- Regulation of actin cytoskeleton - WP523 (Mus musculus)
- ID signaling pathway - WP934 (Pan troglodytes)
- ID signaling pathway - WP397 (Rattus norvegicus)
- ID signaling pathway - WP1168 (Canis familiaris)
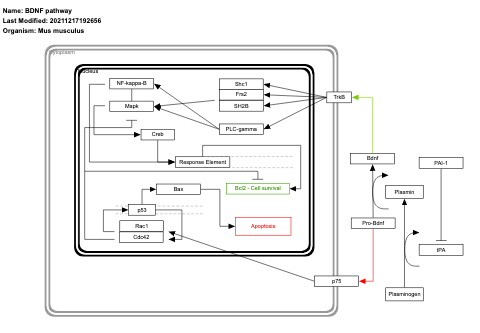
- BDNF pathway - WP2152 (Mus musculus)
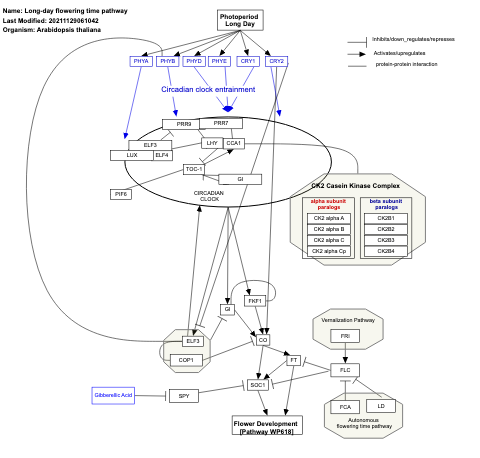
- Long-day flowering time pathway - WP622 (Arabidopsis thaliana)
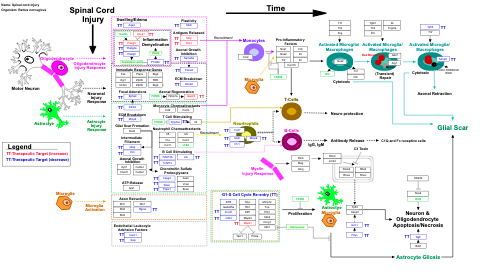
- Spinal cord injury - WP2433 (Rattus norvegicus)
- Nod-like receptor (NLR) signaling pathway - WP799 (Gallus gallus)
- Role of carnosine in muscle contraction - WP4486 (Homo sapiens)
- Host-pathogen interaction of human coronaviruses - MAPK signaling - WP4877 (Homo sapiens)
- miR-222 in exercise-induced cardiac growth - WP2928 (Mus musculus)
- Flowering time pathway - WP2603 (Beta vulgaris)
- Nodal signaling pathway - WP341 (Danio rerio)
- Flavonoid pathway - WP3620 (Arabidopsis thaliana)
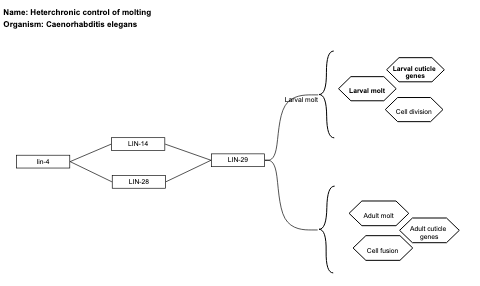
- Heterchronic control of molting - WP2357 (Caenorhabditis elegans)
- FGF signaling pathway - WP152 (Danio rerio)
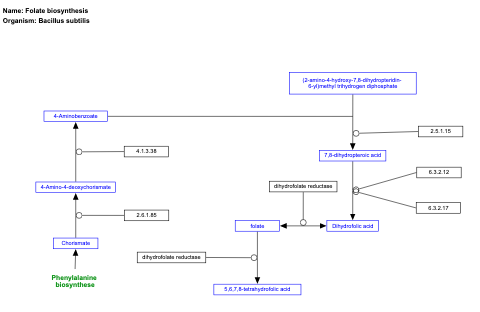
- Folate biosynthesis - WP2360 (Bacillus subtilis)
- Wnt signaling pathway and pluripotency - WP723 (Mus musculus)
- EGFR1 signaling pathway - WP572 (Mus musculus)
- Exercise-induced circadian regulation - WP544 (Mus musculus)
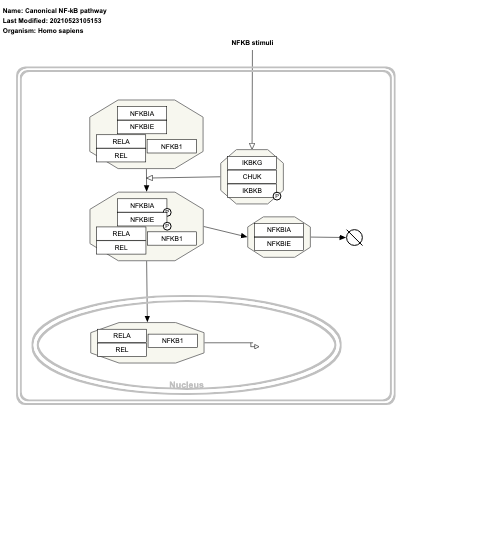
- Canonical NF-kB pathway - WP4562 (Homo sapiens)
- Kit receptor signaling pathway - WP407 (Mus musculus)
- IL-6 signaling pathway - WP387 (Mus musculus)
- Aryl hydrocarbon receptor pathway - WP3205 (Bos taurus)
- Delta-Notch signaling pathway - WP265 (Mus musculus)
- Neural crest differentiation - WP2074 (Mus musculus)
- GPCRs, orphan - WP1398 (Mus musculus)
- Endochondral ossification - WP1270 (Mus musculus)
- Delta-Notch signaling pathway - WP1180 (Canis familiaris)
- Hedgehog signaling pathway - WP116 (Mus musculus)
- GPCRs, class C metabotropic glutamate, pheromone - WP501 (Homo sapiens)
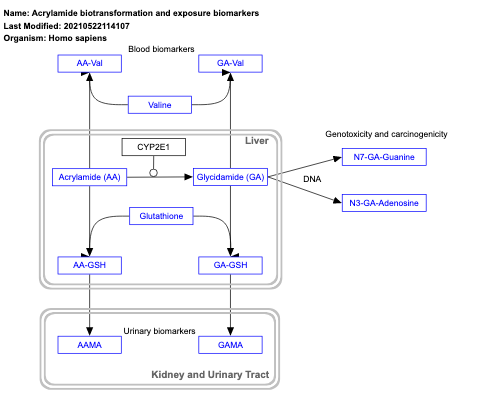
- Acrylamide biotransformation and exposure biomarkers - WP4233 (Homo sapiens)
- Tgif disruption of Shh signaling - WP3674 (Homo sapiens)
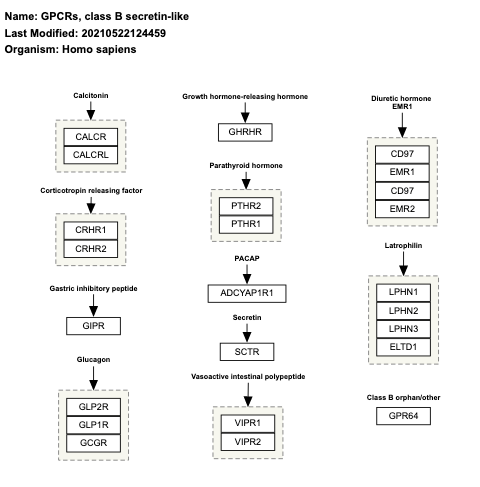
- GPCRs, class B secretin-like - WP334 (Homo sapiens)
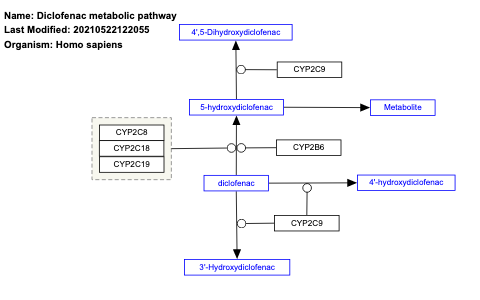
- Diclofenac metabolic pathway - WP2491 (Homo sapiens)
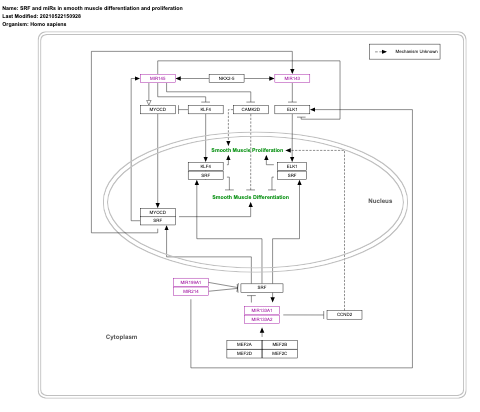
- SRF and miRs in smooth muscle differentiation and proliferation - WP1991 (Homo sapiens)
- Cytokines and inflammatory response - WP997 (Bos taurus)
- Methylation pathways - WP992 (Bos taurus)
- Nuclear receptors in lipid metabolism and toxicity - WP981 (Bos taurus)
- Delta-Notch signaling pathway - WP946 (Pan troglodytes)
- Hedgehog signaling pathway - WP905 (Pan troglodytes)
- Kit receptor signaling pathway - WP886 (Pan troglodytes)
- EGFR1 signaling pathway - WP860 (Pan troglodytes)
- Fatty acid omega-oxidation - WP852 (Pan troglodytes)
- Globo sphingolipid metabolism - WP3278 (Bos taurus)
- Aryl hydrocarbon receptor - WP3226 (Bos taurus)
- ATM signaling pathway - WP3221 (Bos taurus)
- AMPK signaling - WP3154 (Bos taurus)
- Folate-alcohol and cancer pathway - WP3120 (Bos taurus)
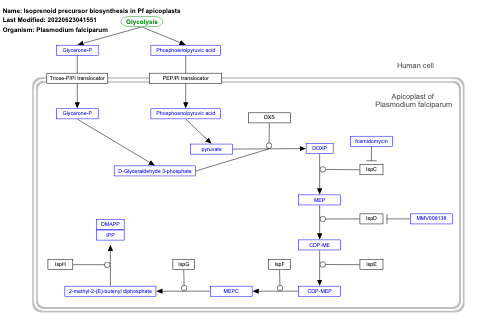
- Isoprenoid precursor biosynthesis in Pf apicoplasts - WP2918 (Plasmodium falciparum)
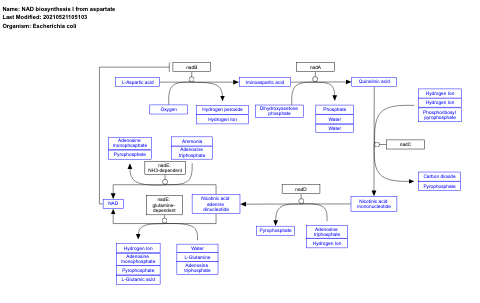
- NAD biosynthesis I from aspartate - WP2484 (Escherichia coli)
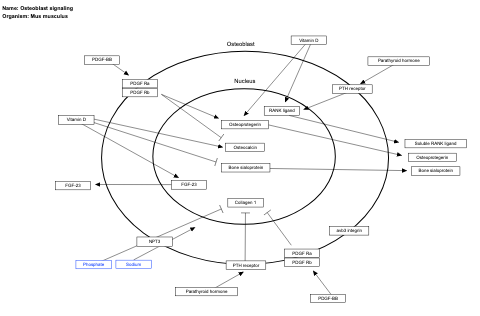
- Osteoblast signaling - WP238 (Mus musculus)
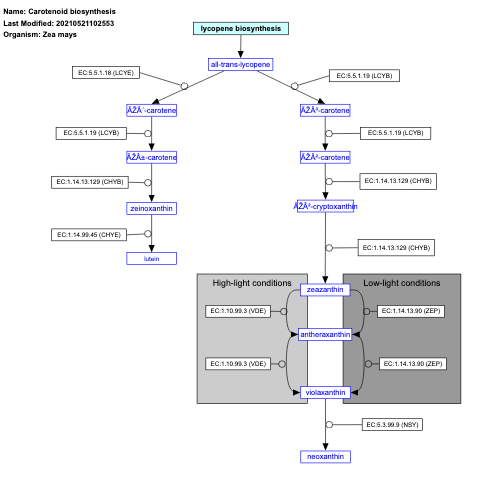
- Carotenoid biosynthesis - WP2205 (Zea mays)
- Ganglio sphingolipid metabolism - WP1423 (Homo sapiens)
- Statin pathway - WP1157 (Canis familiaris)
- Hedgehog signaling pathway - WP1141 (Canis familiaris)
- Kit receptor signaling pathway - WP1121 (Canis familiaris)
- Cytokines and inflammatory response - WP1113 (Canis familiaris)
- EGFR1 signaling pathway - WP1096 (Canis familiaris)
- Endochondral ossification - WP1065 (Bos taurus)
- MAPK signaling pathway - WP510 (Saccharomyces cerevisiae)
- Inhibitor of DNA binding (ID) signaling pathway - WP815 (Gallus gallus)
- Hedgehog signaling pathway - WP790 (Gallus gallus)
- Kit receptor signaling pathway - WP774 (Gallus gallus)
- Triacylglycerol biosynthesis - WP627 (Arabidopsis thaliana)
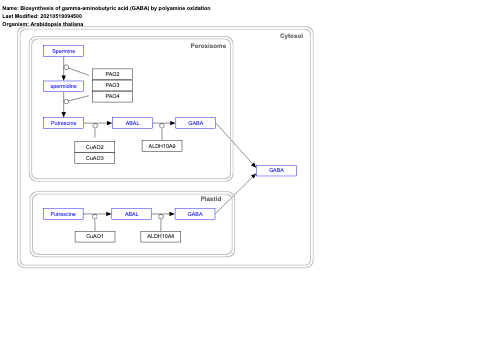
- Biosynthesis of gamma-aminobutyric acid (GABA) by polyamine oxidation - WP4232 (Arabidopsis thaliana)
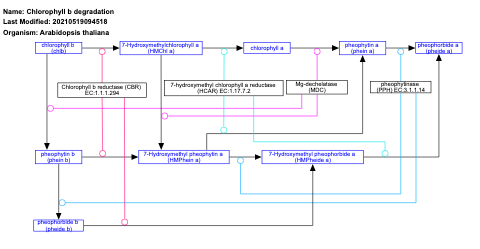
- Chlorophyll b degradation - WP2230 (Arabidopsis thaliana)
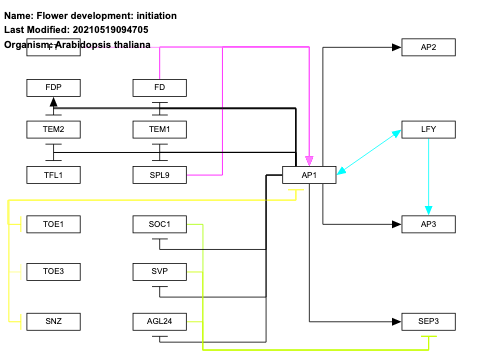
- Flower development: initiation - WP2108 (Arabidopsis thaliana)
- Delta-Notch signaling pathway - WP827 (Gallus gallus)
- EGFR1 signaling pathway - WP752 (Gallus gallus)
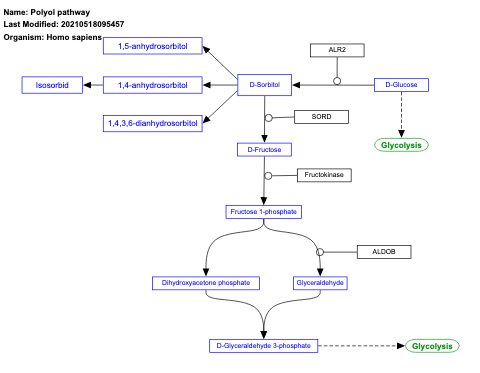
- Polyol pathway - WP690 (Homo sapiens)
- Nod-like receptor (NLR) signaling pathway - WP914 (Pan troglodytes)
- Hedgehog signaling pathway - WP574 (Rattus norvegicus)
- Hedgehog signaling pathway - WP492 (Drosophila melanogaster)
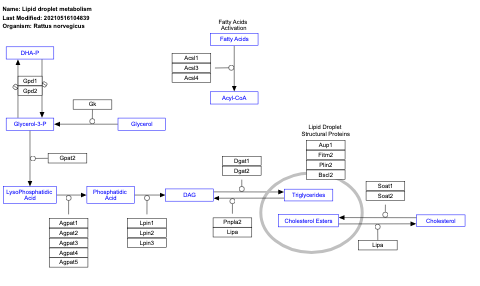
- Lipid droplet metabolism - WP3901 (Rattus norvegicus)
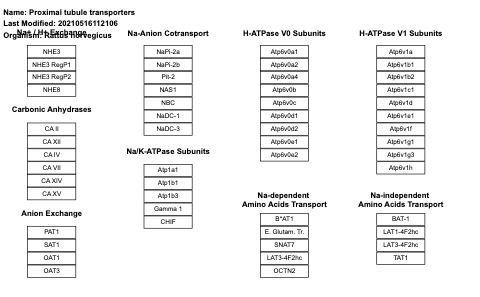
- Proximal tubule transporters - WP3881 (Rattus norvegicus)
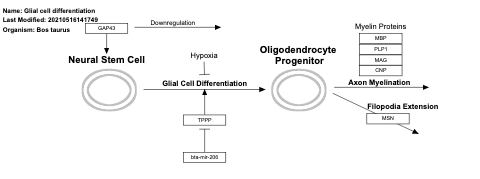
- Glial cell differentiation - WP3203 (Bos taurus)
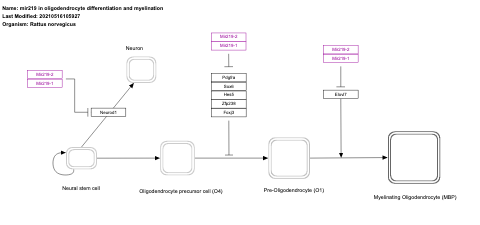
- mir219 in oligodendrocyte differentiation and myelination - WP2811 (Rattus norvegicus)
- Kit receptor signaling pathway - WP147 (Rattus norvegicus)
- Glycolysis and gluconeogenesis - WP1356 (Danio rerio)
- Estrogen signaling - WP1330 (Danio rerio)
- Biogenic amine synthesis - WP522 (Mus musculus)
- Microglia pathogen phagocytosis pathway - WP3626 (Mus musculus)
- MicroRNAs in cardiomyocyte hypertrophy - WP1560 (Mus musculus)
- p38 MAPK signaling pathway - WP1154 (Canis familiaris)
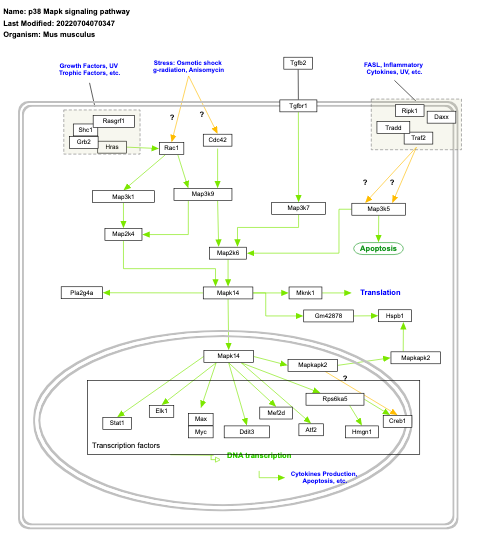
- p38 Mapk signaling pathway - WP350 (Mus musculus)
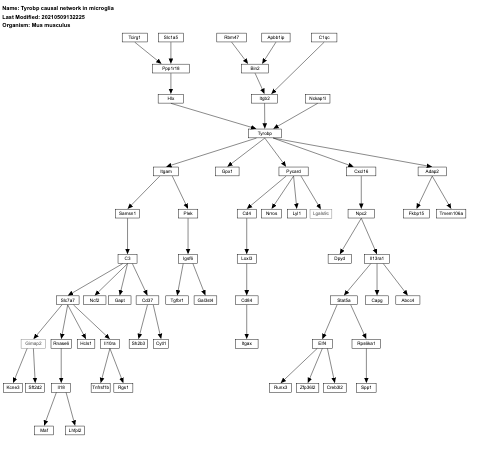
- Tyrobp causal network in microglia - WP3625 (Mus musculus)
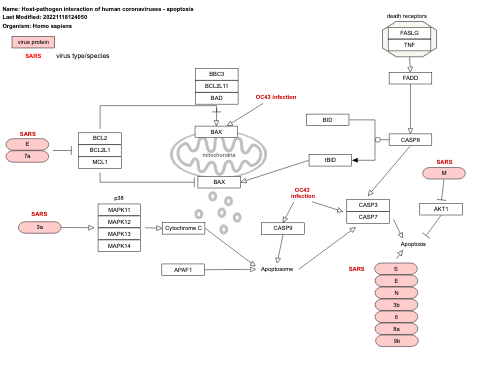
- Host-pathogen interaction of human coronaviruses - apoptosis - WP4864 (Homo sapiens)
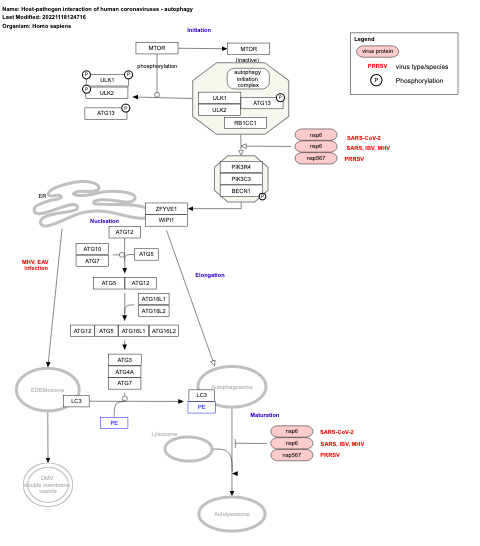
- Host-pathogen interaction of human coronaviruses - autophagy - WP4863 (Homo sapiens)
- Toll-like receptor signaling - WP88 (Mus musculus)
- Wnt Signaling Pathway and Pluripotency - WP1344 (Danio rerio)
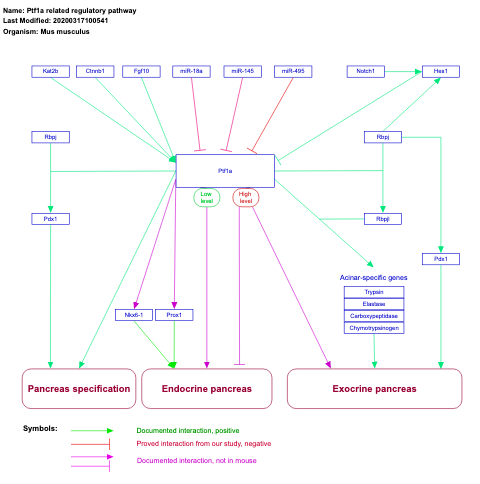
- Ptf1a related regulatory pathway - WP201 (Mus musculus)
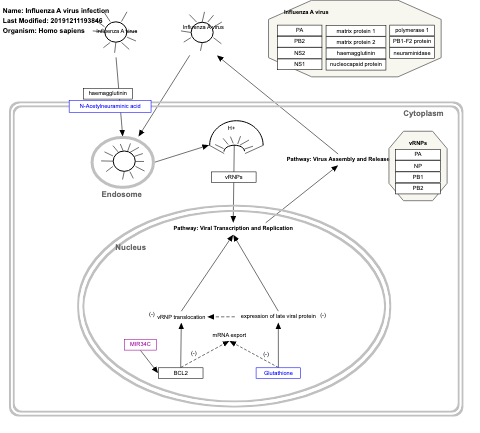
- Influenza A virus infection - WP1438 (Homo sapiens)
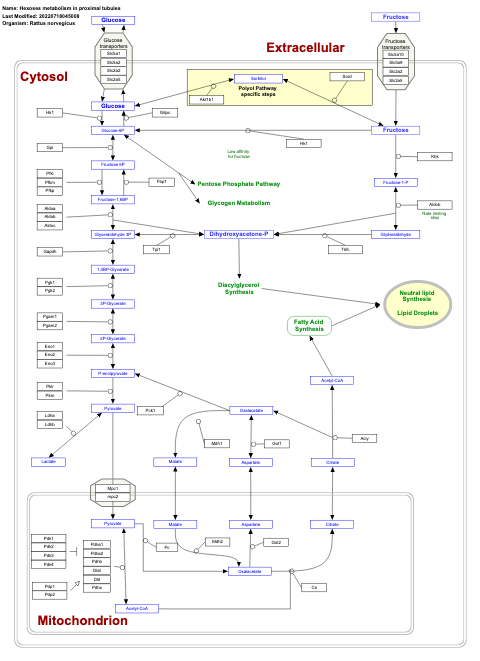
- Hexoses metabolism in proximal tubules - WP3916 (Rattus norvegicus)
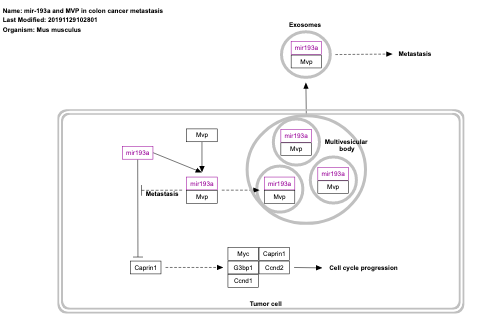
- mir-193a and MVP in colon cancer metastasis - WP3979 (Mus musculus)
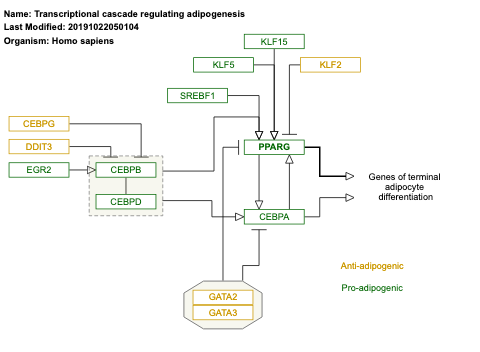
- Transcriptional cascade regulating adipogenesis - WP4211 (Homo sapiens)
- Gonadotropin-releasing hormone (GnRH) signaling - WP2901 (Bos taurus)
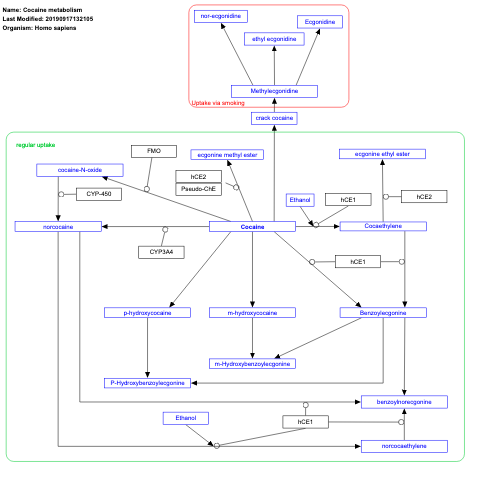
- Cocaine metabolism - WP2826 (Homo sapiens)
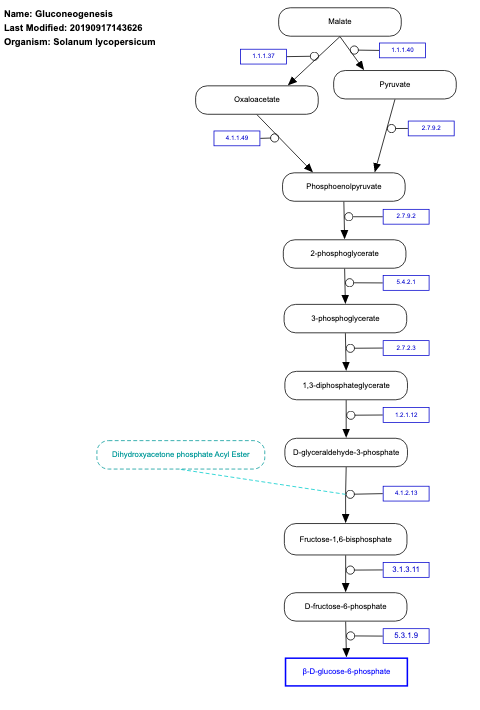
- Gluconeogenesis - WP2630 (Solanum lycopersicum)
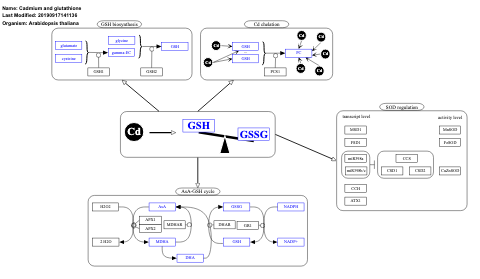
- Cadmium and glutathione - WP2579 (Arabidopsis thaliana)
- PPAR signaling pathway - WP2316 (Mus musculus)
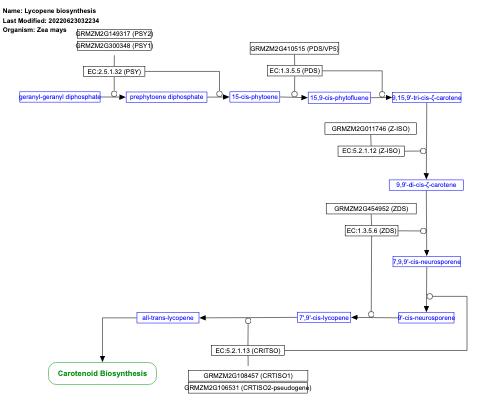
- Lycopene biosynthesis - WP2204 (Zea mays)
- Estrogen signaling pathway - WP986 (Bos taurus)
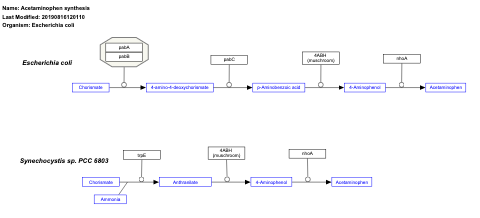
- Acetaminophen synthesis - WP2886 (Escherichia coli)
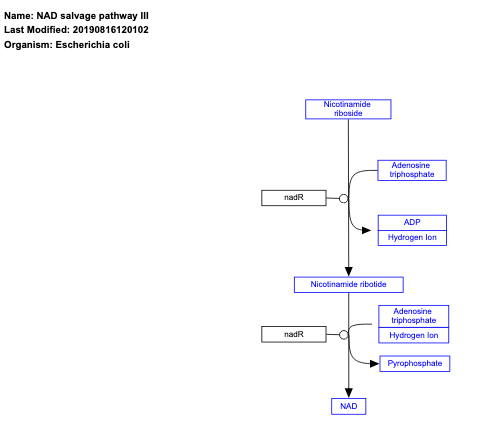
- NAD salvage pathway III - WP2488 (Escherichia coli)
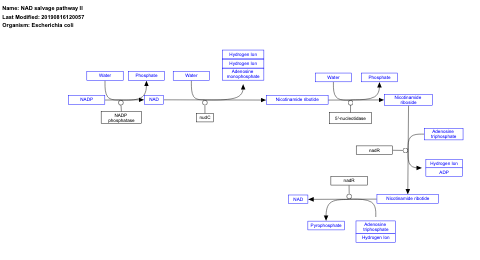
- NAD salvage pathway II - WP2487 (Escherichia coli)
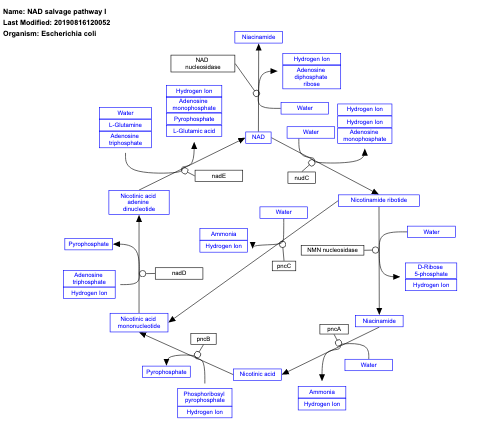
- NAD salvage pathway I - WP2486 (Escherichia coli)
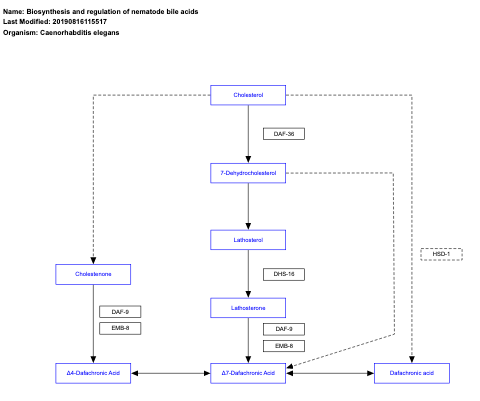
- Biosynthesis and regulation of nematode bile acids - WP2293 (Caenorhabditis elegans)
- Hfe effect on hepcidin production - WP3924 (Homo sapiens)
- Apoptosis - WP1254 (Mus musculus)
- Brain derived neurotrophic factor - WP2148 (Rattus norvegicus)
- Hfe effect on hepcidin production - WP3673 (Mus musculus)
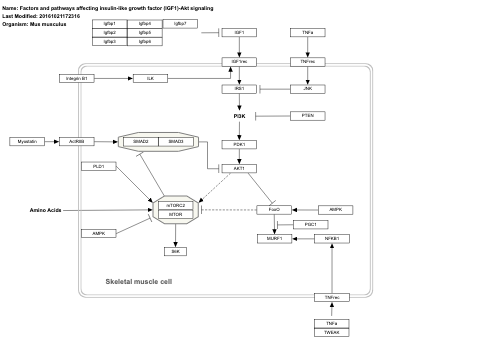
- Factors and pathways affecting insulin-like growth factor (IGF1)-Akt signaling - WP3675 (Mus musculus)
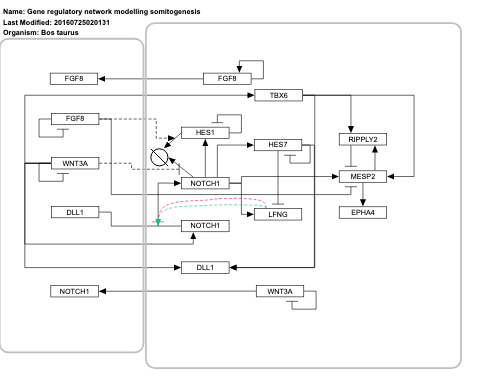
- Gene regulatory network modelling somitogenesis - WP3216 (Bos taurus)
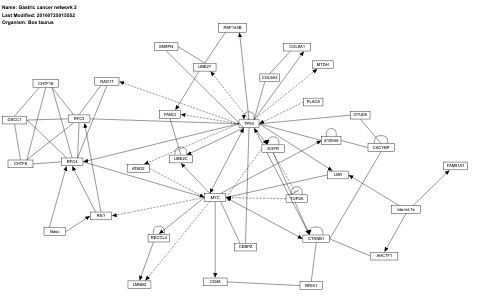
- Gastric cancer network 2 - WP3139 (Bos taurus)
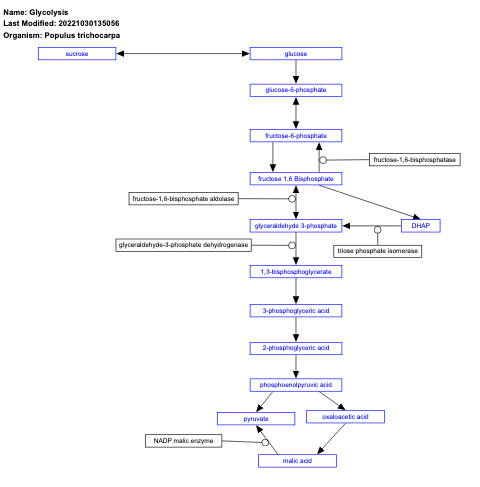
- Glycolysis - WP2862 (Populus trichocarpa)
- Gene regulatory network modelling somitogenesis - WP2852 (Mus musculus)
- Type II interferon signaling (IFNG) - WP1253 (Mus musculus)