Welcome to the COVID-19 Pathway community page!
This community fouses on curating the molecular mechanisms of COVID-19. It is part of a larger community effort called the COVID-19 Disease Map. After an initial comment in 2020 in Scientific Data Ostaszewski, et al., a longer paper from the whole community was published in 2021 in Molecular Systems Biology: Ostaszewski, et al.. More recently, the community is working on a paper focused on different analysis and modelling approaches using the COVID-19 Disease Map, currently available on bioXriv Niarakis, et al.
Contact
Alex Pico , Denise Slenter , Egon Willighagen , Friederike Ehrhart , Martina Summer-Kutmon , and Nhung Pham .
Community Pathways
Table FiltersThis community helps to curate 39 pathways:
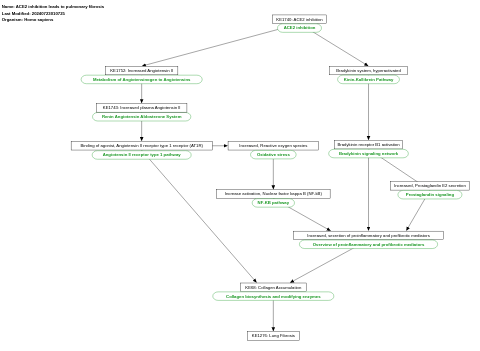
- ACE2 inhibition leads to pulmonary fibrosis - WP5035 (Homo sapiens)
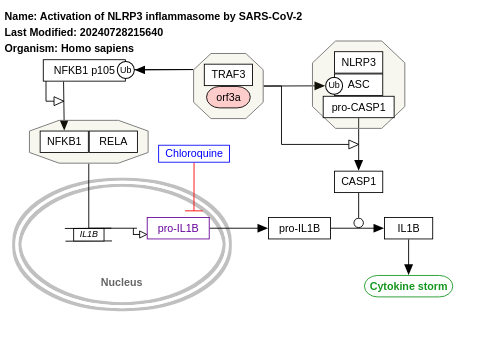
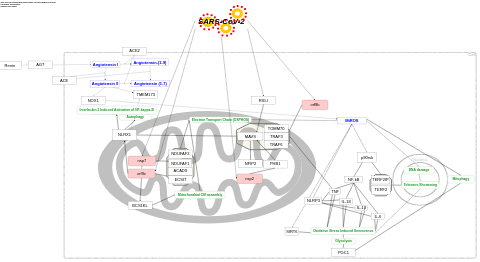
- Activation of NLRP3 inflammasome by SARS-CoV-2 - WP4876 (Homo sapiens)
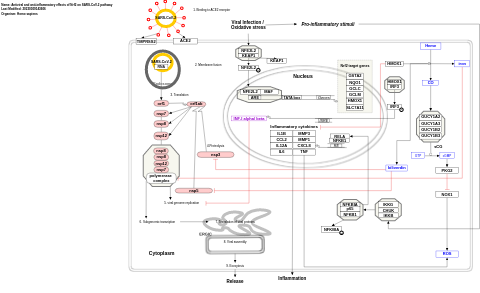
- Antiviral and anti-inflammatory effects of Nrf2 on SARS-CoV-2 pathway - WP5113 (Homo sapiens)
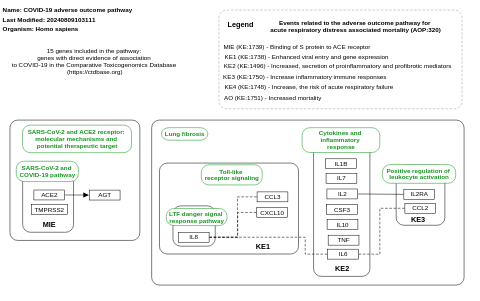
- COVID-19 adverse outcome pathway - WP4891 (Homo sapiens)
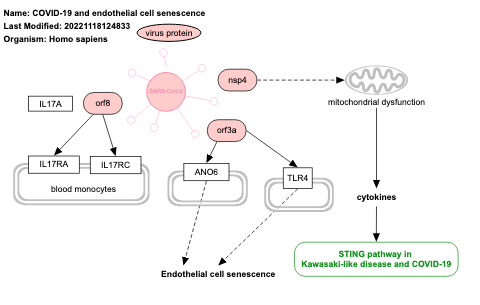
- COVID-19 and endothelial cell senescence - WP5256 (Homo sapiens)
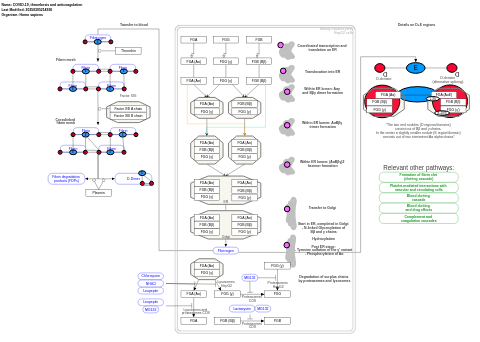
- COVID-19, thrombosis and anticoagulation - WP4927 (Homo sapiens)
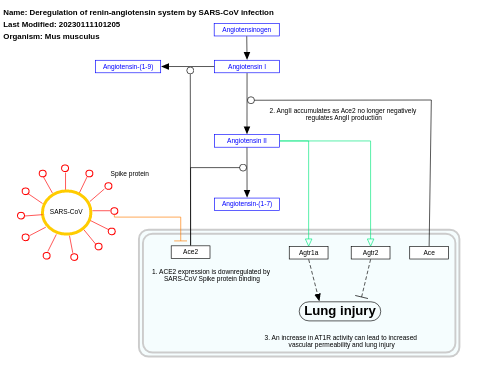
- Deregulation of renin-angiotensin system by SARS-CoV infection - WP4965 (Mus musculus)
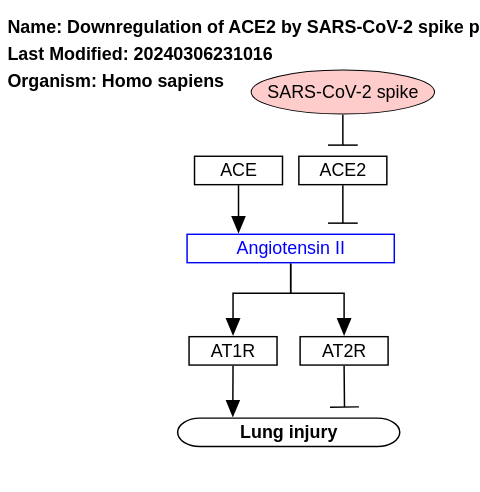
- Downregulation of ACE2 by SARS-CoV-2 spike protein - WP4799 (Homo sapiens)
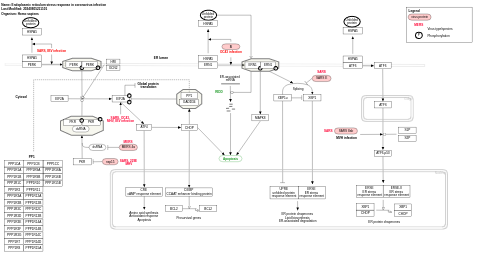
- Endoplasmic reticulum stress response in coronavirus infection - WP4861 (Homo sapiens)
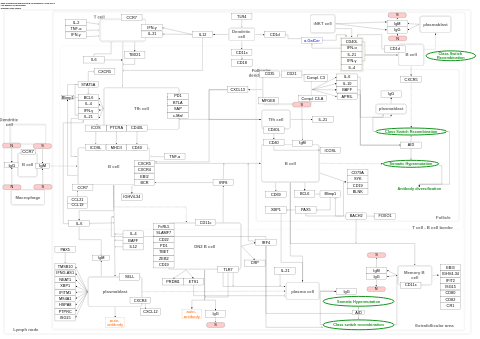
- Extrafollicular and follicular B cell activation by SARS-CoV-2 - WP5218 (Homo sapiens)
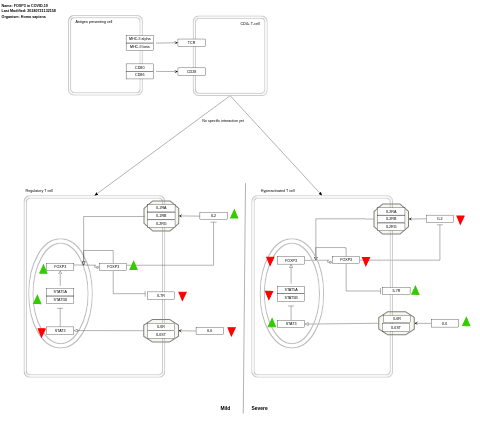
- FOXP3 in COVID-19 - WP5063 (Homo sapiens)
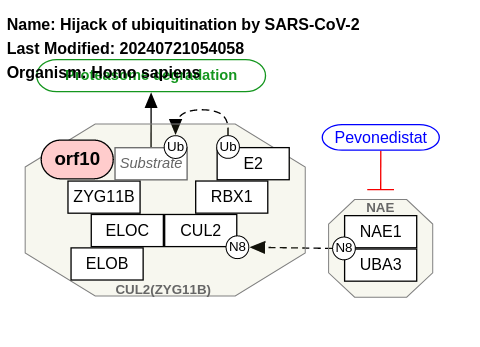
- Hijack of ubiquitination by SARS-CoV-2 - WP4860 (Homo sapiens)
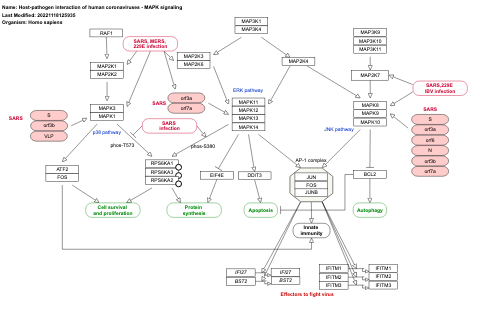
- Host-pathogen interaction of human coronaviruses - MAPK signaling - WP4877 (Homo sapiens)
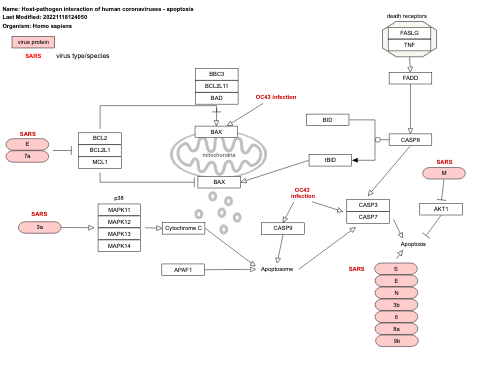
- Host-pathogen interaction of human coronaviruses - apoptosis - WP4864 (Homo sapiens)
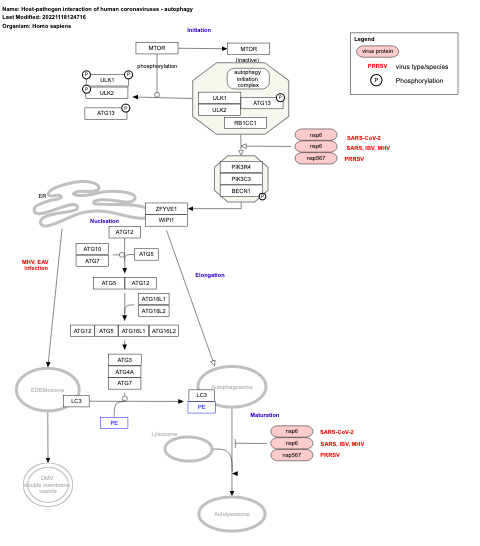
- Host-pathogen interaction of human coronaviruses - autophagy - WP4863 (Homo sapiens)
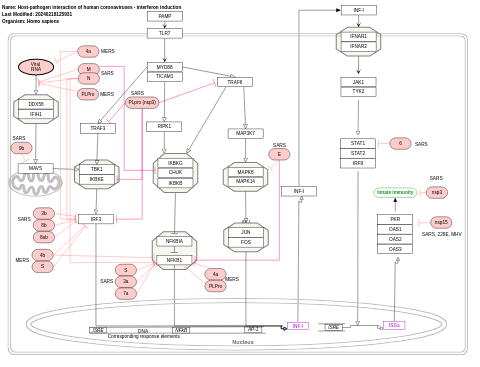
- Host-pathogen interaction of human coronaviruses - interferon induction - WP4880 (Homo sapiens)
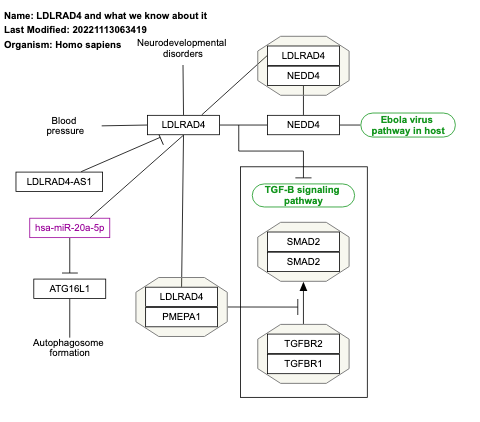
- LDLRAD4 and what we know about it - WP4904 (Homo sapiens)
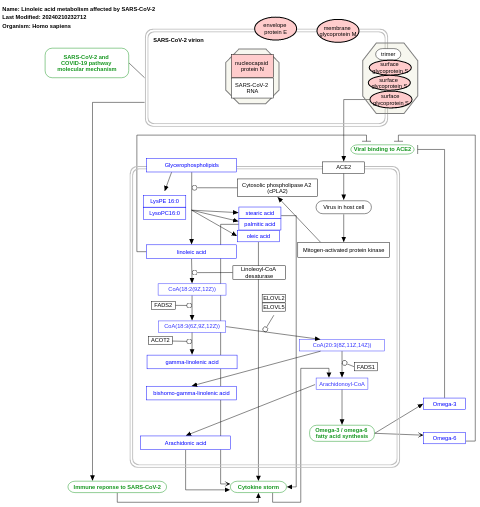
- Linoleic acid metabolism affected by SARS-CoV-2 - WP4853 (Homo sapiens)
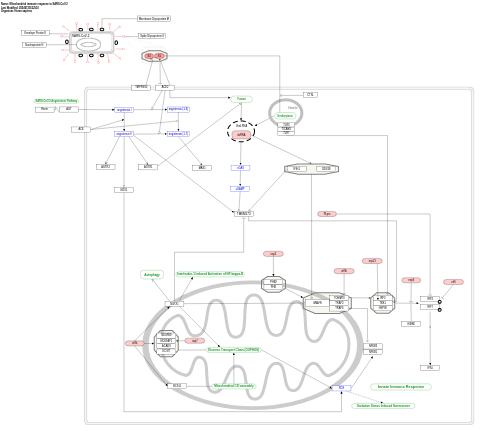
- Mitochondrial immune response to SARS-CoV-2 - WP5038 (Homo sapiens)
- Network map of SARS-CoV-2 signaling - WP5115 (Homo sapiens)
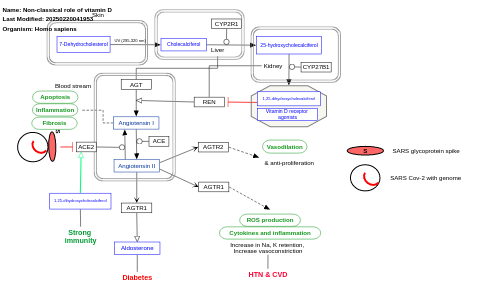
- Non-classical role of vitamin D - WP5133 (Homo sapiens)
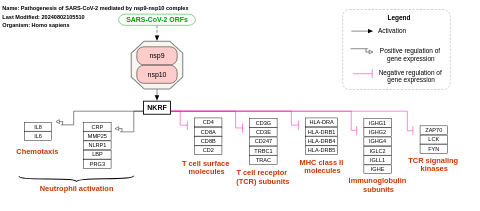
- Pathogenesis of SARS-CoV-2 mediated by nsp9-nsp10 complex - WP4884 (Homo sapiens)
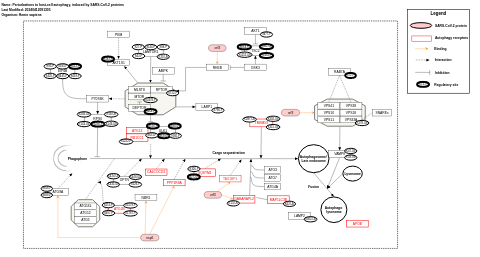
- Perturbations to host-cell autophagy, induced by SARS-CoV-2 proteins - WP4936 (Homo sapiens)
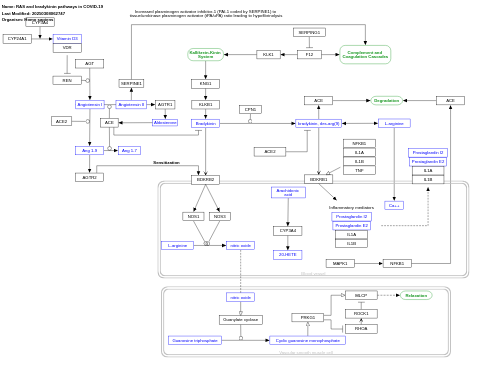
- RAS and bradykinin pathways in COVID-19 - WP4969 (Homo sapiens)
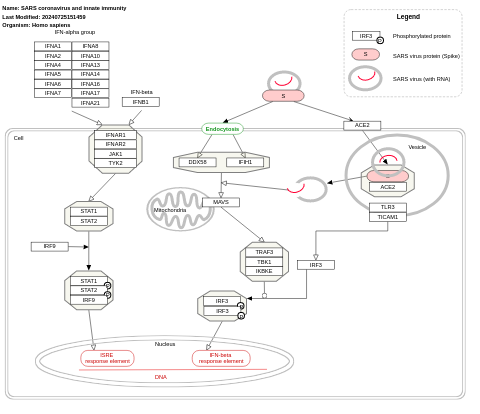
- SARS coronavirus and innate immunity - WP4912 (Homo sapiens)
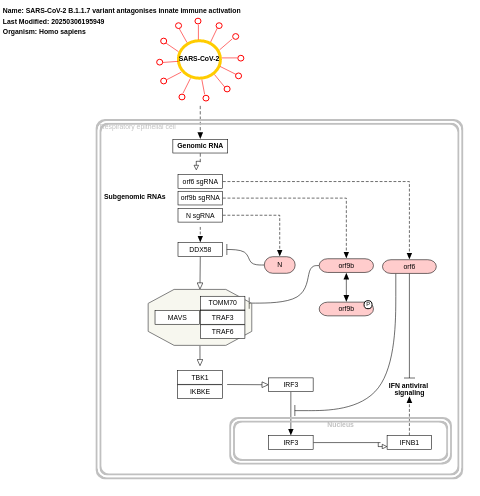
- SARS-CoV-2 B.1.1.7 variant antagonises innate immune activation - WP5116 (Homo sapiens)
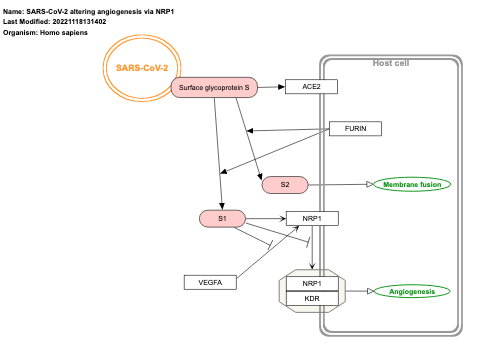
- SARS-CoV-2 altering angiogenesis via NRP1 - WP5065 (Homo sapiens)
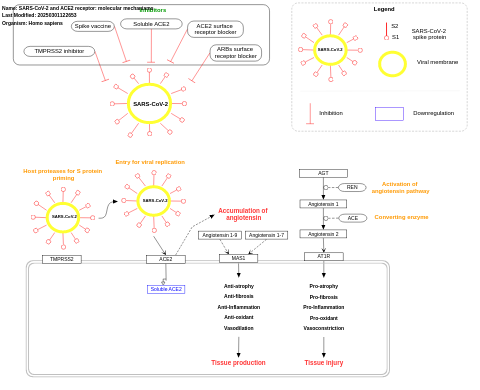
- SARS-CoV-2 and ACE2 receptor: molecular mechanisms - WP4883 (Homo sapiens)
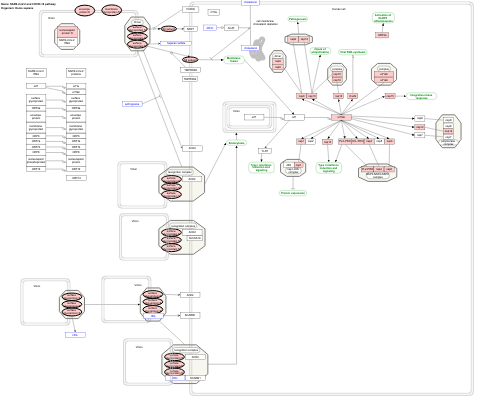
- SARS-CoV-2 and COVID-19 pathway - WP4846 (Homo sapiens)
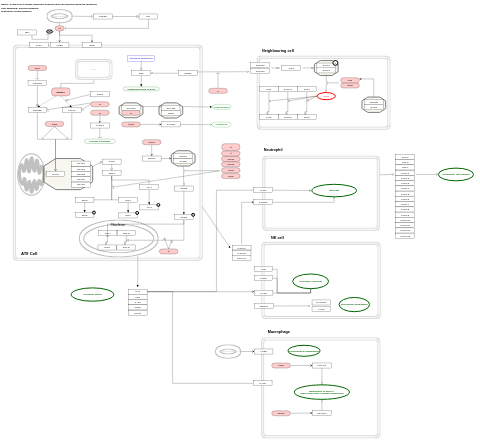
- SARS-CoV-2 innate immunity evasion and cell-specific immune response - WP5039 (Homo sapiens)
- SARS-CoV-2 mitochondrial chronic oxidative stress and endothelial dysfunction - WP5183 (Homo sapiens)
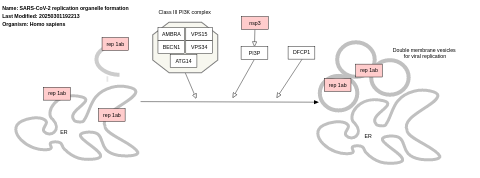
- SARS-CoV-2 replication organelle formation - WP5156 (Homo sapiens)
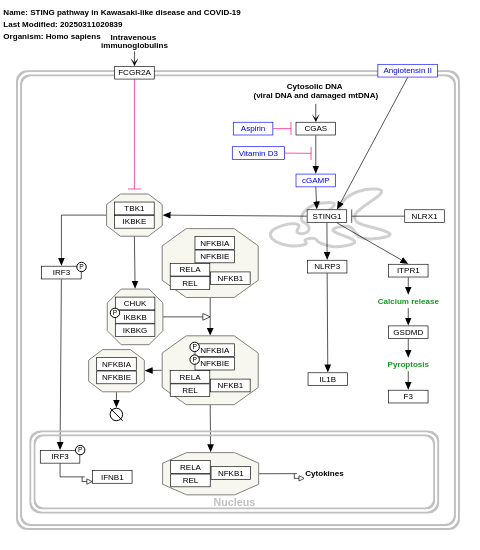
- STING pathway in Kawasaki-like disease and COVID-19 - WP4961 (Homo sapiens)
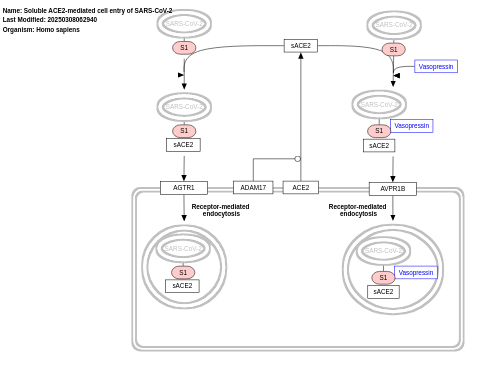
- Soluble ACE2-mediated cell entry of SARS-CoV-2 - WP5076 (Homo sapiens)
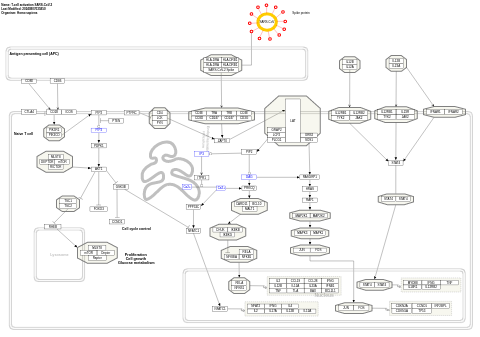
- T-cell activation SARS-CoV-2 - WP5098 (Homo sapiens)
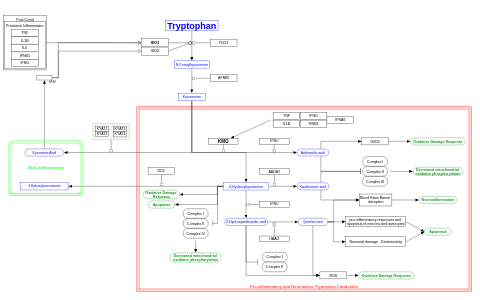
- Tryptophan kynurenine pathway in post-COVID syndrome - WP5549 (Homo sapiens)
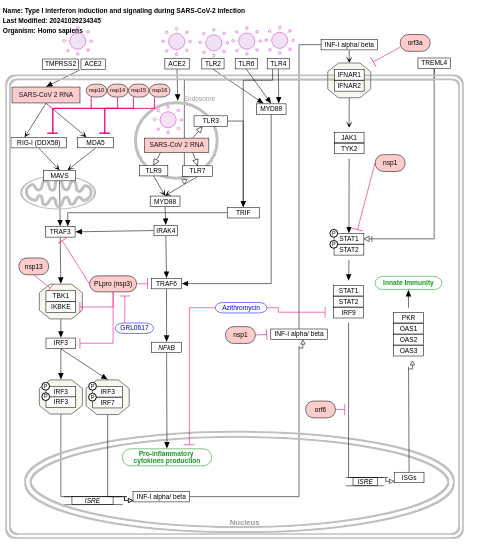
- Type I interferon induction and signaling during SARS-CoV-2 infection - WP4868 (Homo sapiens)
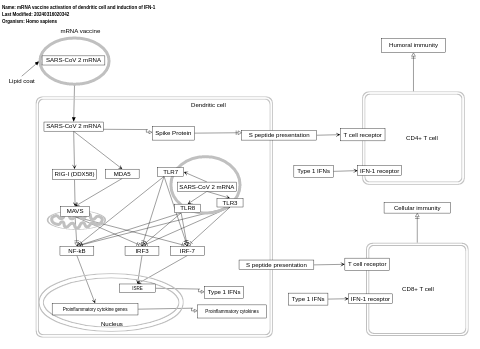
- mRNA vaccine activation of dendritic cell and induction of IFN-1 - WP5187 (Homo sapiens)
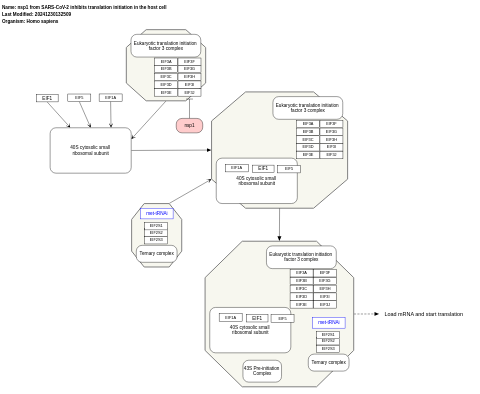
- nsp1 from SARS-CoV-2 inhibits translation initiation in the host cell - WP5027 (Homo sapiens)
How to Contribute
We are helping to coordinate an international effort to build and curate pathway models relevant to the COVID-19 pandemic. If you find or add a pathway at WikiPathways that should be included in this collection, please let us know by contacting Martina Kutmon (mkutmon[AT]gmail.com). Resources to get started:
- Published pathway figures related to COVID-19
- Protein sequence alignments of SARS-CoV-2
- COVID-19 disease map initiative: a broader community of pathway curators working with WikiPathways, Pathway Commons, Reactome, Cell Designer and more.
- Human Coronavirus protein identifiers and SARS-CoV-2 protein identifiers
- PubChem list of the COVID-19 Portal pathways
- KG COVID-19: a knowledge graph aggregating many sources as RDF
To start a discussion about topics relevant to this community, or to browse existing discussions, visit WikiPathways Discussions.
Community Members
Alex Pico , Denise Slenter , Egon Willighagen , Friederike Ehrhart , Martina Summer-Kutmon , and Nhung Pham .
Authors of Community Pathways
Alex Pico , Denise Slenter , Egon Willighagen , Friederike Ehrhart , Martina Summer-Kutmon , Nhung Pham , Marvin Martens , Finterly Hu , Eric Weitz , Rocheliene , Penny Nymark , Chris Evelo , Daniela Digles , Laure-Alix Clerbaux , John Figueira Hasbun , Kristina Hanspers , Jordi Doijen , Amber Koning , Andra Waagmeester , Laurent Winckers , Nuray Talih , Ilja De Wolf , Isabel Wassink , Rex D A B , Matthew Conroy , Cedric Pluis , Lieke Van Den Bogaart , B.P.E. Mennen , Gabriel Couillaud , Claire Billingsley , Kingson ''junsheng Thang'' Luc , Dr. T. S. Keshava Prasad , Susan Coort , Martijn Van De Garde , Lars Willighagen , Hiromasa Ono , Aishwarya Iyer , Humera Fiaz , Lauren J. Dupuis , Simon Vossen , Lena Szabo , Mario Alin Rus , Celine Malyar , Jing Jing Cai , Calvin Classon , Kristof Kirps , and Anna Niarakis .